| 1 | c3ke8A_
|
|
 |
100.0 |
100 |
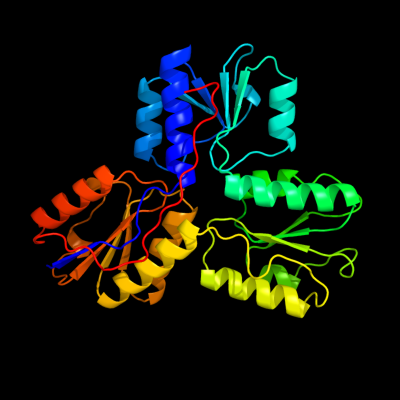
PDB header:oxidoreductase
Chain: A: PDB Molecule:4-hydroxy-3-methylbut-2-enyl diphosphate
PDBTitle: crystal structure of isph:hmbpp-complex
|
| 2 | c3dnfB_
|
|
 |
100.0 |
33 |
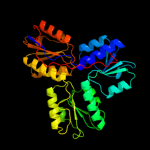
PDB header:oxidoreductase
Chain: B: PDB Molecule:4-hydroxy-3-methylbut-2-enyl diphosphate reductase;
PDBTitle: structure of (e)-4-hydroxy-3-methyl-but-2-enyl diphosphate reductase,2 the terminal enzyme of the non-mevalonate pathway
|
| 3 | c3j09A_
|
|
 |
86.1 |
13 |
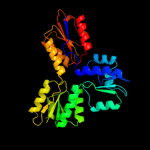
PDB header:hydrolase, metal transport
Chain: A: PDB Molecule:copper-exporting p-type atpase a;
PDBTitle: high resolution helical reconstruction of the bacterial p-type atpase2 copper transporter copa
|
| 4 | d1ma3a_
|
|
 |
84.9 |
15 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Sir2 family of transcriptional regulators |
| 5 | d1m2ka_
|
|
 |
84.7 |
15 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Sir2 family of transcriptional regulators |
| 6 | c3ff4A_
|
|
 |
84.5 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of uncharacterized protein chu_1412
|
| 7 | c3pkiF_
|
|
 |
79.1 |
17 |
PDB header:hydrolase
Chain: F: PDB Molecule:nad-dependent deacetylase sirtuin-6;
PDBTitle: human sirt6 crystal structure in complex with adp ribose
|
| 8 | d1s5pa_
|
|
 |
76.5 |
9 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Sir2 family of transcriptional regulators |
| 9 | c3h5lB_
|
|
 |
76.3 |
16 |
PDB header:transport protein
Chain: B: PDB Molecule:putative branched-chain amino acid abc
PDBTitle: crystal structure of a putative branched-chain amino acid2 abc transporter from silicibacter pomeroyi
|
| 10 | c1kblA_
|
|
 |
75.7 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:pyruvate phosphate dikinase;
PDBTitle: pyruvate phosphate dikinase
|
| 11 | c3j08A_
|
|
 |
75.1 |
12 |
PDB header:hydrolase, metal transport
Chain: A: PDB Molecule:copper-exporting p-type atpase a;
PDBTitle: high resolution helical reconstruction of the bacterial p-type atpase2 copper transporter copa
|
| 12 | d1yc5a1
|
|
 |
74.8 |
19 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Sir2 family of transcriptional regulators |
| 13 | d1j6ua1
|
|
 |
74.5 |
12 |
Fold:MurCD N-terminal domain
Superfamily:MurCD N-terminal domain
Family:MurCD N-terminal domain |
| 14 | c2bg5C_
|
|
 |
72.6 |
17 |
PDB header:transferase
Chain: C: PDB Molecule:phosphoenolpyruvate-protein kinase;
PDBTitle: crystal structure of the phosphoenolpyruvate-binding enzyme2 i-domain from the thermoanaerobacter tengcongensis pep:3 sugar phosphotransferase system (pts)
|
| 15 | c3afoB_
|
|
 |
72.3 |
10 |
PDB header:transferase
Chain: B: PDB Molecule:nadh kinase pos5;
PDBTitle: crystal structure of yeast nadh kinase complexed with nadh
|
| 16 | c2olsA_
|
|
 |
71.4 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoenolpyruvate synthase;
PDBTitle: the crystal structure of the phosphoenolpyruvate synthase from2 neisseria meningitidis
|
| 17 | c3l4eA_
|
|
 |
69.8 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:uncharacterized peptidase lmo0363;
PDBTitle: 1.5a crystal structure of a putative peptidase e protein from listeria2 monocytogenes egd-e
|
| 18 | c3d22A_
|
|
 |
69.8 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin h-type;
PDBTitle: crystal structure of a poplar thioredoxin h mutant,2 pttrxh4c61s
|
| 19 | d1xmta_
|
|
 |
69.7 |
20 |
Fold:Acyl-CoA N-acyltransferases (Nat)
Superfamily:Acyl-CoA N-acyltransferases (Nat)
Family:N-acetyl transferase, NAT |
| 20 | d1s1pa_
|
|
 |
69.0 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 21 | d1fyea_ |
|
not modelled |
68.2 |
17 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Aspartyl dipeptidase PepE |
| 22 | c3rfuC_ |
|
not modelled |
67.1 |
15 |
PDB header:hydrolase, membrane protein
Chain: C: PDB Molecule:copper efflux atpase;
PDBTitle: crystal structure of a copper-transporting pib-type atpase
|
| 23 | d1qo0a_ |
|
not modelled |
65.5 |
16 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
| 24 | c3cumA_ |
|
not modelled |
65.4 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable 3-hydroxyisobutyrate dehydrogenase;
PDBTitle: crystal structure of a possible 3-hydroxyisobutyrate dehydrogenase2 from pseudomonas aeruginosa pao1
|
| 25 | c3h75A_ |
|
not modelled |
65.1 |
10 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:periplasmic sugar-binding domain protein;
PDBTitle: crystal structure of a periplasmic sugar-binding protein from the2 pseudomonas fluorescens
|
| 26 | d1kbla1 |
|
not modelled |
64.2 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate phosphate dikinase, C-terminal domain |
| 27 | d1oj7a_ |
|
not modelled |
63.7 |
10 |
Fold:Dehydroquinate synthase-like
Superfamily:Dehydroquinate synthase-like
Family:Iron-containing alcohol dehydrogenase |
| 28 | d2dlxa1 |
|
not modelled |
62.8 |
11 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:UAS domain |
| 29 | d1oboa_ |
|
not modelled |
62.2 |
7 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Flavodoxin-related |
| 30 | d2j13a1 |
|
not modelled |
61.8 |
12 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycoside hydrolase/deacetylase
Family:NodB-like polysaccharide deacetylase |
| 31 | c3k35D_ |
|
not modelled |
60.0 |
15 |
PDB header:hydrolase
Chain: D: PDB Molecule:nad-dependent deacetylase sirtuin-6;
PDBTitle: crystal structure of human sirt6
|
| 32 | c2iz6A_ |
|
not modelled |
59.3 |
17 |
PDB header:metal transport
Chain: A: PDB Molecule:molybdenum cofactor carrier protein;
PDBTitle: structure of the chlamydomonas rheinhardtii moco carrier2 protein
|
| 33 | c1j6uA_ |
|
not modelled |
59.0 |
15 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramate-alanine ligase murc;
PDBTitle: crystal structure of udp-n-acetylmuramate-alanine ligase2 murc (tm0231) from thermotoga maritima at 2.3 a resolution
|
| 34 | d1e0ta2 |
|
not modelled |
57.9 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 35 | d1pkla2 |
|
not modelled |
57.4 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 36 | c3ej6D_ |
|
not modelled |
57.3 |
20 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:catalase-3;
PDBTitle: neurospora crassa catalase-3 crystal structure
|
| 37 | c2vlvA_ |
|
not modelled |
56.8 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin h isoform 2.;
PDBTitle: crystal structure of barley thioredoxin h isoform 2 in2 partially radiation-reduced state
|
| 38 | d1w5fa1 |
|
not modelled |
56.3 |
23 |
Fold:Tubulin nucleotide-binding domain-like
Superfamily:Tubulin nucleotide-binding domain-like
Family:Tubulin, GTPase domain |
| 39 | c1jzdA_ |
|
not modelled |
55.9 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thiol:disulfide interchange protein dsbc;
PDBTitle: dsbc-dsbdalpha complex
|
| 40 | c3mw8A_ |
|
not modelled |
54.8 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:uroporphyrinogen-iii synthase;
PDBTitle: crystal structure of an uroporphyrinogen-iii synthase (sama_3255) from2 shewanella amazonensis sb2b at 1.65 a resolution
|
| 41 | c2hroA_ |
|
not modelled |
54.4 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoenolpyruvate-protein phosphotransferase;
PDBTitle: structure of the full-lenght enzyme i of the pts system from2 staphylococcus carnosus
|
| 42 | d1ep7a_ |
|
not modelled |
54.3 |
15 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Thioltransferase |
| 43 | d1ag9a_ |
|
not modelled |
54.0 |
10 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Flavodoxin-related |
| 44 | c3snrA_ |
|
not modelled |
53.7 |
16 |
PDB header:transport protein
Chain: A: PDB Molecule:extracellular ligand-binding receptor;
PDBTitle: rpd_1889 protein, an extracellular ligand-binding receptor from2 rhodopseudomonas palustris.
|
| 45 | c3oa3A_ |
|
not modelled |
53.1 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:aldolase;
PDBTitle: crystal structure of a putative deoxyribose-phosphate aldolase from2 coccidioides immitis
|
| 46 | c3s40C_ |
|
not modelled |
52.9 |
15 |
PDB header:transferase
Chain: C: PDB Molecule:diacylglycerol kinase;
PDBTitle: the crystal structure of a diacylglycerol kinases from bacillus2 anthracis str. sterne
|
| 47 | d1ofua1 |
|
not modelled |
52.8 |
23 |
Fold:Tubulin nucleotide-binding domain-like
Superfamily:Tubulin nucleotide-binding domain-like
Family:Tubulin, GTPase domain |
| 48 | c1t00A_ |
|
not modelled |
52.5 |
10 |
PDB header:electron transport
Chain: A: PDB Molecule:thioredoxin;
PDBTitle: the structure of thioredoxin from s. coelicolor
|
| 49 | d1h6za1 |
|
not modelled |
52.3 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate phosphate dikinase, C-terminal domain |
| 50 | d1r57a_ |
|
not modelled |
52.2 |
22 |
Fold:Acyl-CoA N-acyltransferases (Nat)
Superfamily:Acyl-CoA N-acyltransferases (Nat)
Family:N-acetyl transferase, NAT |
| 51 | d2d59a1 |
|
not modelled |
51.6 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
| 52 | d2hk6a1 |
|
not modelled |
51.6 |
17 |
Fold:Chelatase-like
Superfamily:Chelatase
Family:Ferrochelatase |
| 53 | d1vbga1 |
|
not modelled |
51.6 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate phosphate dikinase, C-terminal domain |
| 54 | c1vbhA_ |
|
not modelled |
51.3 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:pyruvate,orthophosphate dikinase;
PDBTitle: pyruvate phosphate dikinase with bound mg-pep from maize
|
| 55 | c3h6hB_ |
|
not modelled |
50.5 |
13 |
PDB header:membrane protein
Chain: B: PDB Molecule:glutamate receptor, ionotropic kainate 2;
PDBTitle: crystal structure of the glur6 amino terminal domain dimer assembly2 mpd form
|
| 56 | c3i09A_ |
|
not modelled |
50.4 |
20 |
PDB header:transport protein
Chain: A: PDB Molecule:periplasmic branched-chain amino acid-binding protein;
PDBTitle: crystal structure of a periplasmic binding protein (bma2936) from2 burkholderia mallei at 1.80 a resolution
|
| 57 | d1yq2a5 |
|
not modelled |
49.7 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 58 | c2h9aA_ |
|
not modelled |
49.2 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:carbon monoxide dehydrogenase corrinoid/iron-
PDBTitle: corrinoid iron-sulfur protein
|
| 59 | c1zxxA_ |
|
not modelled |
49.2 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:6-phosphofructokinase;
PDBTitle: the crystal structure of phosphofructokinase from lactobacillus2 delbrueckii
|
| 60 | c2hwgA_ |
|
not modelled |
49.0 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoenolpyruvate-protein phosphotransferase;
PDBTitle: structure of phosphorylated enzyme i of the2 phosphoenolpyruvate:sugar phosphotransferase system
|
| 61 | d2g17a1 |
|
not modelled |
48.9 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 62 | c2vpiA_ |
|
not modelled |
48.8 |
2 |
PDB header:ligase
Chain: A: PDB Molecule:gmp synthase;
PDBTitle: human gmp synthetase - glutaminase domain
|
| 63 | c1w89E_ |
|
not modelled |
48.6 |
16 |
PDB header:electron transport
Chain: E: PDB Molecule:thioredoxin;
PDBTitle: structure of the reduced form of human thioredoxin 2
|
| 64 | d1j6ua2 |
|
not modelled |
48.5 |
14 |
Fold:MurD-like peptide ligases, peptide-binding domain
Superfamily:MurD-like peptide ligases, peptide-binding domain
Family:MurCDEF C-terminal domain |
| 65 | c1t3bA_ |
|
not modelled |
48.4 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:thiol:disulfide interchange protein dsbc;
PDBTitle: x-ray structure of dsbc from haemophilus influenzae
|
| 66 | d2b4ya1 |
|
not modelled |
48.0 |
19 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Sir2 family of transcriptional regulators |
| 67 | c3pfnB_ |
|
not modelled |
46.1 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:nad kinase;
PDBTitle: crystal structure of human nad kinase
|
| 68 | c3tcoA_ |
|
not modelled |
45.9 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin (trxa-1);
PDBTitle: crystallographic and spectroscopic characterization of sulfolobus2 solfataricus trxa1 provide insights into the determinants of3 thioredoxin fold stability
|
| 69 | c3hutA_ |
|
not modelled |
45.1 |
15 |
PDB header:transport protein
Chain: A: PDB Molecule:putative branched-chain amino acid abc
PDBTitle: crystal structure of a putative branched-chain amino acid2 abc transporter from rhodospirillum rubrum
|
| 70 | d2jfga1 |
|
not modelled |
44.8 |
17 |
Fold:MurCD N-terminal domain
Superfamily:MurCD N-terminal domain
Family:MurCD N-terminal domain |
| 71 | d2je8a5 |
|
not modelled |
44.7 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 72 | d1qwka_ |
|
not modelled |
44.2 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 73 | d1a3xa2 |
|
not modelled |
43.8 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 74 | d1t3ba1 |
|
not modelled |
41.7 |
16 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:DsbC/DsbG C-terminal domain-like |
| 75 | c3k2qA_ |
|
not modelled |
41.0 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:pyrophosphate-dependent phosphofructokinase;
PDBTitle: crystal structure of pyrophosphate-dependent2 phosphofructokinase from marinobacter aquaeolei, northeast3 structural genomics consortium target mqr88
|
| 76 | c3gnjD_ |
|
not modelled |
40.4 |
13 |
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:thioredoxin domain protein;
PDBTitle: the crystal structure of a thioredoxin-related protein from2 desulfitobacterium hafniense dcb
|
| 77 | c3la6P_ |
|
not modelled |
40.0 |
10 |
PDB header:transferase
Chain: P: PDB Molecule:tyrosine-protein kinase wzc;
PDBTitle: octameric kinase domain of the e. coli tyrosine kinase wzc with bound2 adp
|
| 78 | d1j96a_ |
|
not modelled |
39.9 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 79 | c3p9zA_ |
|
not modelled |
39.9 |
13 |
PDB header:ligase
Chain: A: PDB Molecule:uroporphyrinogen iii cosynthase (hemd);
PDBTitle: crystal structure of uroporphyrinogen-iii synthetase from helicobacter2 pylori 26695
|
| 80 | d1jhfa1 |
|
not modelled |
39.8 |
15 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:LexA repressor, N-terminal DNA-binding domain |
| 81 | d1p3da1 |
|
not modelled |
39.8 |
19 |
Fold:MurCD N-terminal domain
Superfamily:MurCD N-terminal domain
Family:MurCD N-terminal domain |
| 82 | c2qsiB_ |
|
not modelled |
39.5 |
13 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative hydrogenase expression/formation protein hupg;
PDBTitle: crystal structure of putative hydrogenase expression/formation protein2 hupg from rhodopseudomonas palustris cga009
|
| 83 | c3n0xA_ |
|
not modelled |
39.1 |
18 |
PDB header:transport protein
Chain: A: PDB Molecule:possible substrate binding protein of abc transporter
PDBTitle: crystal structure of an abc-type branched-chain amino acid transporter2 (rpa4397) from rhodopseudomonas palustris cga009 at 1.50 a resolution
|
| 84 | d1jvna2 |
|
not modelled |
38.9 |
16 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 85 | d1vqof1 |
|
not modelled |
38.7 |
21 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:L30e/L7ae ribosomal proteins |
| 86 | c3canA_ |
|
not modelled |
38.7 |
12 |
PDB header:lyase activator
Chain: A: PDB Molecule:pyruvate-formate lyase-activating enzyme;
PDBTitle: crystal structure of a domain of pyruvate-formate lyase-activating2 enzyme from bacteroides vulgatus atcc 8482
|
| 87 | d1hw6a_ |
|
not modelled |
38.6 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 88 | c1v57A_ |
|
not modelled |
38.6 |
13 |
PDB header:isomerase
Chain: A: PDB Molecule:thiol:disulfide interchange protein dsbg;
PDBTitle: crystal structure of the disulfide bond isomerase dsbg
|
| 89 | d1rlga_ |
|
not modelled |
38.5 |
11 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:L30e/L7ae ribosomal proteins |
| 90 | c3g1wB_ |
|
not modelled |
38.3 |
11 |
PDB header:transport protein
Chain: B: PDB Molecule:sugar abc transporter;
PDBTitle: crystal structure of sugar abc transporter (sugar-binding protein)2 from bacillus halodurans
|
| 91 | d1xwaa_ |
|
not modelled |
38.2 |
14 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Thioltransferase |
| 92 | d1lbqa_ |
|
not modelled |
37.7 |
16 |
Fold:Chelatase-like
Superfamily:Chelatase
Family:Ferrochelatase |
| 93 | c3h7uA_ |
|
not modelled |
37.7 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aldo-keto reductase;
PDBTitle: crystal structure of the plant stress-response enzyme akr4c9
|
| 94 | c3cvjB_ |
|
not modelled |
37.5 |
24 |
PDB header:isomerase
Chain: B: PDB Molecule:putative phosphoheptose isomerase;
PDBTitle: crystal structure of a putative phosphoheptose isomerase (bh3325) from2 bacillus halodurans c-125 at 2.00 a resolution
|
| 95 | c3fn9B_ |
|
not modelled |
37.0 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative beta-galactosidase;
PDBTitle: crystal structure of putative beta-galactosidase from bacteroides2 fragilis
|
| 96 | c3hz4A_ |
|
not modelled |
36.9 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin;
PDBTitle: crystal structure of thioredoxin from methanosarcina mazei
|
| 97 | c2xhzC_ |
|
not modelled |
36.3 |
14 |
PDB header:isomerase
Chain: C: PDB Molecule:arabinose 5-phosphate isomerase;
PDBTitle: probing the active site of the sugar isomerase domain from e. coli2 arabinose-5-phosphate isomerase via x-ray crystallography
|
| 98 | d2f48a1 |
|
not modelled |
35.8 |
18 |
Fold:Phosphofructokinase
Superfamily:Phosphofructokinase
Family:Phosphofructokinase |
| 99 | d4pfka_ |
|
not modelled |
35.4 |
11 |
Fold:Phosphofructokinase
Superfamily:Phosphofructokinase
Family:Phosphofructokinase |
| 100 | c3lkbB_ |
|
not modelled |
35.1 |
15 |
PDB header:transport protein
Chain: B: PDB Molecule:probable branched-chain amino acid abc
PDBTitle: crystal structure of a branched chain amino acid abc2 transporter from thermus thermophilus with bound valine
|
| 101 | c3koxA_ |
|
not modelled |
34.8 |
15 |
PDB header:metal binding protein
Chain: A: PDB Molecule:d-ornithine aminomutase e component;
PDBTitle: crystal structure of ornithine 4,5 aminomutase in complex with 2,4-2 diaminobutyrate (anaerobic)
|
| 102 | d1vp5a_ |
|
not modelled |
34.1 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
| 103 | c3p3wC_ |
|
not modelled |
33.9 |
13 |
PDB header:transport protein
Chain: C: PDB Molecule:glutamate receptor 3;
PDBTitle: structure of a dimeric glua3 n-terminal domain (ntd) at 4.2 a2 resolution
|
| 104 | c3glsC_ |
|
not modelled |
33.7 |
24 |
PDB header:hydrolase
Chain: C: PDB Molecule:nad-dependent deacetylase sirtuin-3,
PDBTitle: crystal structure of human sirt3
|
| 105 | c3mn1B_ |
|
not modelled |
33.5 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:probable yrbi family phosphatase;
PDBTitle: crystal structure of probable yrbi family phosphatase from pseudomonas2 syringae pv.phaseolica 1448a
|
| 106 | d1vhca_ |
|
not modelled |
33.4 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 107 | d1thxa_ |
|
not modelled |
32.9 |
14 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Thioltransferase |
| 108 | c3ngjC_ |
|
not modelled |
32.7 |
14 |
PDB header:lyase
Chain: C: PDB Molecule:deoxyribose-phosphate aldolase;
PDBTitle: crystal structure of a putative deoxyribose-phosphate aldolase from2 entamoeba histolytica
|
| 109 | c3s99A_ |
|
not modelled |
32.7 |
9 |
PDB header:lipid binding protein
Chain: A: PDB Molecule:basic membrane lipoprotein;
PDBTitle: crystal structure of a basic membrane lipoprotein from brucella2 melitensis, iodide soak
|
| 110 | c2r8zC_ |
|
not modelled |
32.7 |
9 |
PDB header:hydrolase
Chain: C: PDB Molecule:3-deoxy-d-manno-octulosonate 8-phosphate phosphatase;
PDBTitle: crystal structure of yrbi phosphatase from escherichia coli in complex2 with a phosphate and a calcium ion
|
| 111 | c3p2yA_ |
|
not modelled |
32.6 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alanine dehydrogenase/pyridine nucleotide transhydrogenase;
PDBTitle: crystal structure of alanine dehydrogenase/pyridine nucleotide2 transhydrogenase from mycobacterium smegmatis
|
| 112 | c3cmgA_ |
|
not modelled |
32.4 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative beta-galactosidase;
PDBTitle: crystal structure of putative beta-galactosidase from bacteroides2 fragilis
|
| 113 | d1nria_ |
|
not modelled |
32.0 |
15 |
Fold:SIS domain
Superfamily:SIS domain
Family:mono-SIS domain |
| 114 | c1nriA_ |
|
not modelled |
32.0 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein hi0754;
PDBTitle: crystal structure of putative phosphosugar isomerase hi0754 from2 haemophilus influenzae
|
| 115 | d1pfka_ |
|
not modelled |
31.9 |
14 |
Fold:Phosphofructokinase
Superfamily:Phosphofructokinase
Family:Phosphofructokinase |
| 116 | c1h6zA_ |
|
not modelled |
31.7 |
9 |
PDB header:transferase
Chain: A: PDB Molecule:pyruvate phosphate dikinase;
PDBTitle: 3.0 a resolution crystal structure of glycosomal pyruvate2 phosphate dikinase from trypanosoma brucei
|
| 117 | c3cyvA_ |
|
not modelled |
31.6 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: crystal structure of uroporphyrinogen decarboxylase from2 shigella flexineri: new insights into its catalytic3 mechanism
|
| 118 | d1z2wa1 |
|
not modelled |
31.4 |
14 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:YfcE-like |
| 119 | d1o5za1 |
|
not modelled |
31.3 |
23 |
Fold:MurD-like peptide ligases, peptide-binding domain
Superfamily:MurD-like peptide ligases, peptide-binding domain
Family:Folylpolyglutamate synthetase, C-terminal domain |
| 120 | d2alra_ |
|
not modelled |
31.3 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |