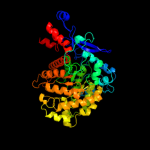
| 1 | c3epmB_ |
|
 |
100.0 |
67 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:thiamine biosynthesis protein thic;
PDBTitle: crystal structure of caulobacter crescentus thic
|
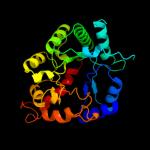
| 2 | c3pnzD_ |
|
 |
95.3 |
13 |
PDB header:hydrolase
Chain: D: PDB Molecule:phosphotriesterase family protein;
PDBTitle: crystal structure of the lactonase lmo2620 from listeria monocytogenes
|
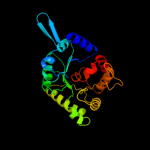
| 3 | d1ad1a_ |
|
 |
93.9 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
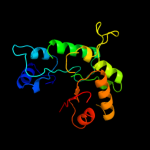
| 4 | d1ub3a_ |
|
 |
91.1 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 5 | c2hjpA_ |
|
 |
90.7 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:phosphonopyruvate hydrolase;
PDBTitle: crystal structure of phosphonopyruvate hydrolase complex with2 phosphonopyruvate and mg++
|
| 6 | d1h1ya_ |
|
 |
90.3 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
| 7 | c3qc3B_ |
|
 |
90.3 |
18 |
PDB header:isomerase
Chain: B: PDB Molecule:d-ribulose-5-phosphate-3-epimerase;
PDBTitle: crystal structure of a d-ribulose-5-phosphate-3-epimerase (np_954699)2 from homo sapiens at 2.20 a resolution
|
| 8 | d1mzha_ |
|
 |
90.1 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 9 | c1tx2A_ |
|
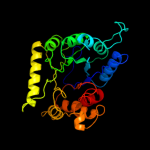 |
89.8 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:dhps, dihydropteroate synthase;
PDBTitle: dihydropteroate synthetase, with bound inhibitor manic, from bacillus2 anthracis
|
| 10 | d1tx2a_ |
|
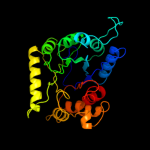 |
89.8 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 11 | c2zvrA_ |
|
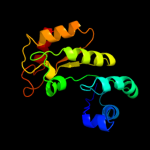 |
89.5 |
11 |
PDB header:isomerase
Chain: A: PDB Molecule:uncharacterized protein tm_0416;
PDBTitle: crystal structure of a d-tagatose 3-epimerase-related2 protein from thermotoga maritima
|
| 12 | d3bofa1 |
|
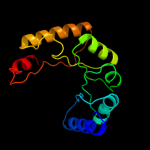 |
89.0 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Methyltetrahydrofolate-utiluzing methyltransferases |
| 13 | c3db2C_ |
|
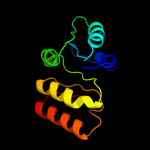 |
87.4 |
18 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative nadph-dependent oxidoreductase;
PDBTitle: crystal structure of a putative nadph-dependent oxidoreductase2 (dhaf_2064) from desulfitobacterium hafniense dcb-2 at 1.70 a3 resolution
|
| 14 | c2vefB_ |
|
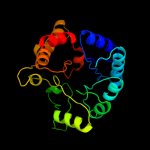 |
87.2 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:dihydropteroate synthase;
PDBTitle: dihydropteroate synthase from streptococcus pneumoniae
|
| 15 | d1ajza_ |
|
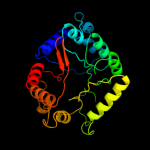 |
86.0 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 16 | d2flia1 |
|
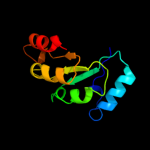 |
85.5 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
| 17 | c3bolB_ |
|
 |
85.2 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:5-methyltetrahydrofolate s-homocysteine
PDBTitle: cobalamin-dependent methionine synthase (1-566) from2 thermotoga maritima complexed with zn2+
|
| 18 | c2o7qA_ |
|
 |
84.4 |
24 |
PDB header:oxidoreductase,transferase
Chain: A: PDB Molecule:bifunctional 3-dehydroquinate dehydratase/shikimate
PDBTitle: crystal structure of the a. thaliana dhq-dehydroshikimate-sdh-2 shikimate-nadp(h)
|
| 19 | d2zdra2 |
|
 |
83.8 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:NeuB-like |
| 20 | d2q02a1 |
|
 |
83.6 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:IolI-like |
| 21 | d1sfla_ |
|
not modelled |
83.4 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 22 | d1i60a_ |
|
not modelled |
83.0 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:IolI-like |
| 23 | c3k13A_ |
|
not modelled |
82.3 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:5-methyltetrahydrofolate-homocysteine methyltransferase;
PDBTitle: structure of the pterin-binding domain metr of 5-2 methyltetrahydrofolate-homocysteine methyltransferase from3 bacteroides thetaiotaomicron
|
| 24 | c2bdqA_ |
|
not modelled |
81.1 |
14 |
PDB header:metal transport
Chain: A: PDB Molecule:copper homeostasis protein cutc;
PDBTitle: crystal structure of the putative copper homeostasis2 protein cutc from streptococcus agalactiae, northeast3 strucural genomics target sar15.
|
| 25 | d1o0ya_ |
|
not modelled |
80.3 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 26 | c3ih1A_ |
|
not modelled |
80.2 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:methylisocitrate lyase;
PDBTitle: crystal structure of carboxyvinyl-carboxyphosphonate phosphorylmutase2 from bacillus anthracis
|
| 27 | d1a3xa2 |
|
not modelled |
80.1 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 28 | c3d0cB_ |
|
not modelled |
80.0 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 oceanobacillus iheyensis at 1.9 a resolution
|
| 29 | c2y0fD_ |
|
not modelled |
79.8 |
19 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase;
PDBTitle: structure of gcpe (ispg) from thermus thermophilus hb27
|
| 30 | c3eooL_ |
|
not modelled |
79.8 |
15 |
PDB header:lyase
Chain: L: PDB Molecule:methylisocitrate lyase;
PDBTitle: 2.9a crystal structure of methyl-isocitrate lyase from2 burkholderia pseudomallei
|
| 31 | c3q2kB_ |
|
not modelled |
79.8 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of the wlba dehydrogenase from bordetella pertussis2 in complex with nadh and udp-glcnaca
|
| 32 | c1zlpA_ |
|
not modelled |
79.3 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:petal death protein;
PDBTitle: petal death protein psr132 with cysteine-linked glutaraldehyde forming2 a thiohemiacetal adduct
|
| 33 | c2c46B_ |
|
not modelled |
79.2 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:mrna capping enzyme;
PDBTitle: crystal structure of the human rna guanylyltransferase and2 5'-phosphatase
|
| 34 | c3k2gA_ |
|
not modelled |
79.2 |
16 |
PDB header:resiniferatoxin binding protein
Chain: A: PDB Molecule:resiniferatoxin-binding, phosphotriesterase-
PDBTitle: crystal structure of a resiniferatoxin-binding protein from2 rhodobacter sphaeroides
|
| 35 | c3ngjC_ |
|
not modelled |
78.5 |
17 |
PDB header:lyase
Chain: C: PDB Molecule:deoxyribose-phosphate aldolase;
PDBTitle: crystal structure of a putative deoxyribose-phosphate aldolase from2 entamoeba histolytica
|
| 36 | c3rbvA_ |
|
not modelled |
77.5 |
17 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:sugar 3-ketoreductase;
PDBTitle: crystal structure of kijd10, a 3-ketoreductase from actinomadura2 kijaniata incomplex with nadp
|
| 37 | c2glxD_ |
|
not modelled |
77.3 |
16 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:1,5-anhydro-d-fructose reductase;
PDBTitle: crystal structure analysis of bacterial 1,5-af reductase
|
| 38 | c2vc7A_ |
|
not modelled |
76.6 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:aryldialkylphosphatase;
PDBTitle: structural basis for natural lactonase and promiscuous2 phosphotriesterase activities
|
| 39 | d1rxda_ |
|
not modelled |
76.6 |
10 |
Fold:(Phosphotyrosine protein) phosphatases II
Superfamily:(Phosphotyrosine protein) phosphatases II
Family:Dual specificity phosphatase-like |
| 40 | c2hg4A_ |
|
not modelled |
76.3 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:6-deoxyerythronolide b synthase;
PDBTitle: structure of the ketosynthase-acyltransferase didomain of module 52 from debs.
|
| 41 | c1vliA_ |
|
not modelled |
75.7 |
17 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:spore coat polysaccharide biosynthesis protein spse;
PDBTitle: crystal structure of spore coat polysaccharide biosynthesis protein2 spse (bsu37870) from bacillus subtilis at 2.38 a resolution
|
| 42 | c2yciX_ |
|
not modelled |
75.7 |
16 |
PDB header:transferase
Chain: X: PDB Molecule:5-methyltetrahydrofolate corrinoid/iron sulfur protein
PDBTitle: methyltransferase native
|
| 43 | d1tqja_ |
|
not modelled |
75.2 |
27 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
| 44 | d1ccwb_ |
|
not modelled |
74.6 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Cobalamin (vitamin B12)-dependent enzymes
Family:Glutamate mutase, large subunit |
| 45 | c3oa3A_ |
|
not modelled |
74.0 |
23 |
PDB header:lyase
Chain: A: PDB Molecule:aldolase;
PDBTitle: crystal structure of a putative deoxyribose-phosphate aldolase from2 coccidioides immitis
|
| 46 | d2qtva4 |
|
not modelled |
72.6 |
23 |
Fold:Gelsolin-like
Superfamily:C-terminal, gelsolin-like domain of Sec23/24
Family:C-terminal, gelsolin-like domain of Sec23/24 |
| 47 | c3l9cA_ |
|
not modelled |
71.6 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: the crystal structure of smu.777 from streptococcus mutans ua159
|
| 48 | c3ngfA_ |
|
not modelled |
71.2 |
10 |
PDB header:isomerase
Chain: A: PDB Molecule:ap endonuclease, family 2;
PDBTitle: crystal structure of ap endonuclease, family 2 from brucella2 melitensis
|
| 49 | c3ng3A_ |
|
not modelled |
71.0 |
23 |
PDB header:lyase
Chain: A: PDB Molecule:deoxyribose-phosphate aldolase;
PDBTitle: crystal structure of deoxyribose phosphate aldolase from mycobacterium2 avium 104 in a schiff base with an unknown aldehyde
|
| 50 | d1f6ya_ |
|
not modelled |
71.0 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Methyltetrahydrofolate-utiluzing methyltransferases |
| 51 | c3cqkB_ |
|
not modelled |
70.6 |
12 |
PDB header:isomerase
Chain: B: PDB Molecule:l-ribulose-5-phosphate 3-epimerase ulae;
PDBTitle: crystal structure of l-xylulose-5-phosphate 3-epimerase ulae (form b)2 complex with zn2+ and sulfate
|
| 52 | d1qt1a_ |
|
not modelled |
70.3 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 53 | c3b0vD_ |
|
not modelled |
70.1 |
26 |
PDB header:oxidoreductase/rna
Chain: D: PDB Molecule:trna-dihydrouridine synthase;
PDBTitle: trna-dihydrouridine synthase from thermus thermophilus in complex with2 trna
|
| 54 | c3noeA_ |
|
not modelled |
70.0 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from pseudomonas2 aeruginosa
|
| 55 | d1ryda1 |
|
not modelled |
70.0 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 56 | c3hf3A_ |
|
not modelled |
69.8 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:chromate reductase;
PDBTitle: old yellow enzyme from thermus scotoductus sa-01
|
| 57 | c3euwB_ |
|
not modelled |
69.5 |
10 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:myo-inositol dehydrogenase;
PDBTitle: crystal structure of a myo-inositol dehydrogenase from corynebacterium2 glutamicum atcc 13032
|
| 58 | d1vbga1 |
|
not modelled |
69.3 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate phosphate dikinase, C-terminal domain |
| 59 | c3gr7A_ |
|
not modelled |
69.2 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadph dehydrogenase;
PDBTitle: structure of oye from geobacillus kaustophilus, hexagonal2 crystal form
|
| 60 | c3khdC_ |
|
not modelled |
68.2 |
19 |
PDB header:transferase
Chain: C: PDB Molecule:pyruvate kinase;
PDBTitle: crystal structure of pff1300w.
|
| 61 | c2dzaA_ |
|
not modelled |
67.9 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: crystal structure of dihydropteroate synthase from thermus2 thermophilus hb8 in complex with 4-aminobenzoate
|
| 62 | c3nt5B_ |
|
not modelled |
67.9 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:inositol 2-dehydrogenase/d-chiro-inositol 3-dehydrogenase;
PDBTitle: crystal structure of myo-inositol dehydrogenase from bacillus subtilis2 with bound cofactor and product inosose
|
| 63 | c3tr9A_ |
|
not modelled |
67.4 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: structure of a dihydropteroate synthase (folp) in complex with pteroic2 acid from coxiella burnetii
|
| 64 | c3fokH_ |
|
not modelled |
67.4 |
10 |
PDB header:structural genomics, unknown function
Chain: H: PDB Molecule:uncharacterized protein cgl0159;
PDBTitle: crystal structure of cgl0159 from corynebacterium2 glutamicum (brevibacterium flavum). northeast structural3 genomics target cgr115
|
| 65 | d1vhna_ |
|
not modelled |
67.2 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 66 | c3daqB_ |
|
not modelled |
67.0 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from methicillin-2 resistant staphylococcus aureus
|
| 67 | c3e96B_ |
|
not modelled |
67.0 |
20 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 bacillus clausii
|
| 68 | c3noyA_ |
|
not modelled |
65.9 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase;
PDBTitle: crystal structure of ispg (gcpe)
|
| 69 | d1h6za1 |
|
not modelled |
65.9 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate phosphate dikinase, C-terminal domain |
| 70 | d1ojxa_ |
|
not modelled |
65.5 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 71 | c3e18A_ |
|
not modelled |
65.4 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of nad-binding protein from listeria innocua
|
| 72 | c1ps9A_ |
|
not modelled |
65.2 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2,4-dienoyl-coa reductase;
PDBTitle: the crystal structure and reaction mechanism of e. coli 2,4-2 dienoyl coa reductase
|
| 73 | d1rpxa_ |
|
not modelled |
64.1 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
| 74 | c3rgqA_ |
|
not modelled |
64.0 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:protein-tyrosine phosphatase mitochondrial 1;
PDBTitle: crystal structure of ptpmt1 in complex with pi(5)p
|
| 75 | c3fd8A_ |
|
not modelled |
63.4 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of an oxidoreductase from enterococcus2 faecalis
|
| 76 | c3kruC_ |
|
not modelled |
63.4 |
27 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:nadh:flavin oxidoreductase/nadh oxidase;
PDBTitle: crystal structure of the thermostable old yellow enzyme from2 thermoanaerobacter pseudethanolicus e39
|
| 77 | d1gkra2 |
|
not modelled |
62.7 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Metallo-dependent hydrolases
Family:Hydantoinase (dihydropyrimidinase), catalytic domain |
| 78 | c3fluD_ |
|
not modelled |
62.5 |
14 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from the pathogen2 neisseria meningitidis
|
| 79 | c1ofgF_ |
|
not modelled |
62.4 |
18 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:glucose-fructose oxidoreductase;
PDBTitle: glucose-fructose oxidoreductase
|
| 80 | c1h6dL_ |
|
not modelled |
62.4 |
18 |
PDB header:protein translocation
Chain: L: PDB Molecule:precursor form of glucose-fructose
PDBTitle: oxidized precursor form of glucose-fructose oxidoreductase2 from zymomonas mobilis complexed with glycerol
|
| 81 | d1pkla2 |
|
not modelled |
62.1 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 82 | c2q4eB_ |
|
not modelled |
61.6 |
10 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable oxidoreductase at4g09670;
PDBTitle: ensemble refinement of the protein crystal structure of gene product2 from arabidopsis thaliana at4g09670
|
| 83 | d1xbta1 |
|
not modelled |
61.5 |
19 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Type II thymidine kinase |
| 84 | c3nsnA_ |
|
not modelled |
60.6 |
30 |
PDB header:hydrolase
Chain: A: PDB Molecule:n-acetylglucosaminidase;
PDBTitle: crystal structure of insect beta-n-acetyl-d-hexosaminidase ofhex12 complexed with tmg-chitotriomycin
|
| 85 | c3l5aA_ |
|
not modelled |
58.9 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadh/flavin oxidoreductase/nadh oxidase;
PDBTitle: crystal structure of a probable nadh-dependent flavin oxidoreductase2 from staphylococcus aureus
|
| 86 | c3ceaA_ |
|
not modelled |
58.7 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:myo-inositol 2-dehydrogenase;
PDBTitle: crystal structure of myo-inositol 2-dehydrogenase (np_786804.1) from2 lactobacillus plantarum at 2.40 a resolution
|
| 87 | d1vjia_ |
|
not modelled |
58.4 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 88 | c3p6lA_ |
|
not modelled |
58.4 |
12 |
PDB header:isomerase
Chain: A: PDB Molecule:sugar phosphate isomerase/epimerase;
PDBTitle: crystal structure of a sugar phosphate isomerase/epimerase (bdi_1903)2 from parabacteroides distasonis atcc 8503 at 1.85 a resolution
|
| 89 | c2yxgD_ |
|
not modelled |
57.9 |
18 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihyrodipicolinate synthase (dapa)
|
| 90 | d2glka1 |
|
not modelled |
57.9 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 91 | d1n7ka_ |
|
not modelled |
57.8 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 92 | d2icsa2 |
|
not modelled |
57.6 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Metallo-dependent hydrolases
Family:Adenine deaminase-like |
| 93 | d1kbla1 |
|
not modelled |
57.6 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate phosphate dikinase, C-terminal domain |
| 94 | d1vcva1 |
|
not modelled |
57.1 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 95 | d1eyea_ |
|
not modelled |
56.6 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 96 | c1vrdA_ |
|
not modelled |
56.6 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine-5'-monophosphate dehydrogenase;
PDBTitle: crystal structure of inosine-5'-monophosphate dehydrogenase (tm1347)2 from thermotoga maritima at 2.18 a resolution
|
| 97 | c3obkH_ |
|
not modelled |
56.1 |
20 |
PDB header:lyase
Chain: H: PDB Molecule:delta-aminolevulinic acid dehydratase;
PDBTitle: crystal structure of delta-aminolevulinic acid dehydratase2 (porphobilinogen synthase) from toxoplasma gondii me49 in complex3 with the reaction product porphobilinogen
|
| 98 | c1zfjA_ |
|
not modelled |
55.2 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine monophosphate dehydrogenase;
PDBTitle: inosine monophosphate dehydrogenase (impdh; ec 1.1.1.205) from2 streptococcus pyogenes
|
| 99 | c3qyqC_ |
|
not modelled |
55.1 |
15 |
PDB header:lyase
Chain: C: PDB Molecule:deoxyribose-phosphate aldolase, putative;
PDBTitle: 1.8 angstrom resolution crystal structure of a putative deoxyribose-2 phosphate aldolase from toxoplasma gondii me49
|
| 100 | c3js3C_ |
|
not modelled |
54.9 |
15 |
PDB header:lyase
Chain: C: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: crystal structure of type i 3-dehydroquinate dehydratase (arod) from2 clostridium difficile with covalent reaction intermediate
|
| 101 | c3moiA_ |
|
not modelled |
54.7 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable dehydrogenase;
PDBTitle: the crystal structure of the putative dehydrogenase from bordetella2 bronchiseptica rb50
|
| 102 | c3gkaB_ |
|
not modelled |
54.5 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:n-ethylmaleimide reductase;
PDBTitle: crystal structure of n-ethylmaleimidine reductase from2 burkholderia pseudomallei
|
| 103 | d1xkya1 |
|
not modelled |
54.5 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 104 | d1muwa_ |
|
not modelled |
54.2 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 105 | c3s4oB_ |
|
not modelled |
54.2 |
13 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:protein tyrosine phosphatase-like protein;
PDBTitle: protein tyrosine phosphatase (putative) from leishmania major
|
| 106 | c2imgA_ |
|
not modelled |
53.3 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:dual specificity protein phosphatase 23;
PDBTitle: crystal structure of dual specificity protein phosphatase2 23 from homo sapiens in complex with ligand malate ion
|
| 107 | d1a53a_ |
|
not modelled |
53.0 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 108 | c2x5eA_ |
|
not modelled |
52.5 |
25 |
PDB header:unknown function
Chain: A: PDB Molecule:upf0271 protein pa4511;
PDBTitle: crystal structure of the hypothetical protein pa4511 from2 pseudomonas aeruginosa
|
| 109 | c2qjhH_ |
|
not modelled |
52.4 |
22 |
PDB header:lyase
Chain: H: PDB Molecule:putative aldolase mj0400;
PDBTitle: m. jannaschii adh synthase covalently bound to2 dihydroxyacetone phosphate
|
| 110 | c3eb2A_ |
|
not modelled |
52.0 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:putative dihydrodipicolinate synthetase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 2.0a resolution
|
| 111 | d1fpza_ |
|
not modelled |
51.2 |
17 |
Fold:(Phosphotyrosine protein) phosphatases II
Superfamily:(Phosphotyrosine protein) phosphatases II
Family:Dual specificity phosphatase-like |
| 112 | c3dxeB_ |
|
not modelled |
51.2 |
20 |
PDB header:protein binding
Chain: B: PDB Molecule:amyloid beta a4 protein;
PDBTitle: crystal structure of the intracellular domain of human app2 (t668a mutant) in complex with fe65-ptb2
|
| 113 | c1xuzA_ |
|
not modelled |
50.2 |
18 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:polysialic acid capsule biosynthesis protein siac;
PDBTitle: crystal structure analysis of sialic acid synthase (neub)from2 neisseria meningitidis, bound to mn2+, phosphoenolpyruvate, and n-3 acetyl mannosaminitol
|
| 114 | c2oczA_ |
|
not modelled |
49.9 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: the structure of a putative 3-dehydroquinate dehydratase from2 streptococcus pyogenes.
|
| 115 | c2o48X_ |
|
not modelled |
49.1 |
14 |
PDB header:oxidoreductase
Chain: X: PDB Molecule:dimeric dihydrodiol dehydrogenase;
PDBTitle: crystal structure of mammalian dimeric dihydrodiol dehydrogenase
|
| 116 | d1k77a_ |
|
not modelled |
48.9 |
8 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Hypothetical protein YgbM (EC1530) |
| 117 | c2elrA_ |
|
not modelled |
48.9 |
58 |
PDB header:transcription
Chain: A: PDB Molecule:zinc finger protein 406;
PDBTitle: solution structure of the 15th c2h2 zinc finger of human2 zinc finger protein 406
|
| 118 | d1vc4a_ |
|
not modelled |
48.8 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 119 | d1d3ga_ |
|
not modelled |
48.1 |
25 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 120 | c3dx5A_ |
|
not modelled |
47.2 |
9 |
PDB header:lyase
Chain: A: PDB Molecule:uncharacterized protein asbf;
PDBTitle: crystal structure of the probable 3-dhs dehydratase asbf involved in2 the petrobactin synthesis from bacillus anthracis
|

