| 1 | c1zfnA_ |
|
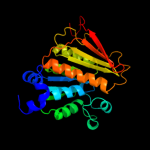 |
100.0 |
100 |
PDB header:transferase
Chain: A: PDB Molecule:adenylyltransferase thif;
PDBTitle: structural analysis of escherichia coli thif
|
| 2 | d1jw9b_ |
|
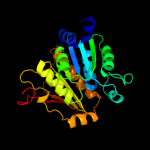 |
100.0 |
46 |
Fold:Activating enzymes of the ubiquitin-like proteins
Superfamily:Activating enzymes of the ubiquitin-like proteins
Family:Molybdenum cofactor biosynthesis protein MoeB |
| 3 | c3h9gA_ |
|
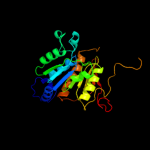 |
100.0 |
27 |
PDB header:transferase/antibiotic
Chain: A: PDB Molecule:mccb protein;
PDBTitle: crystal structure of e. coli mccb + mcca-n7isoasn
|
| 4 | d1yovb1 |
|
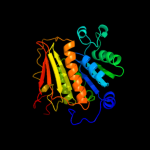 |
100.0 |
27 |
Fold:Activating enzymes of the ubiquitin-like proteins
Superfamily:Activating enzymes of the ubiquitin-like proteins
Family:Ubiquitin activating enzymes (UBA) |
| 5 | c3gznB_ |
|
 |
100.0 |
25 |
PDB header:protein binding/ligase
Chain: B: PDB Molecule:nedd8-activating enzyme e1 catalytic subunit;
PDBTitle: structure of nedd8-activating enzyme in complex with nedd82 and mln4924
|
| 6 | c3vh3A_ |
|
 |
100.0 |
21 |
PDB header:metal binding protein/protein transport
Chain: A: PDB Molecule:ubiquitin-like modifier-activating enzyme atg7;
PDBTitle: crystal structure of atg7ctd-atg8 complex
|
| 7 | c1y8qA_ |
|
 |
100.0 |
24 |
PDB header:ligase
Chain: A: PDB Molecule:ubiquitin-like 1 activating enzyme e1a;
PDBTitle: sumo e1 activating enzyme sae1-sae2-mg-atp complex
|
| 8 | c2nvuB_ |
|
 |
100.0 |
30 |
PDB header:protein turnover, ligase
Chain: B: PDB Molecule:maltose binding protein/nedd8-activating enzyme
PDBTitle: structure of appbp1-uba3~nedd8-nedd8-mgatp-ubc12(c111a), a2 trapped ubiquitin-like protein activation complex
|
| 9 | c3kydB_ |
|
 |
100.0 |
28 |
PDB header:ligase
Chain: B: PDB Molecule:sumo-activating enzyme subunit 2;
PDBTitle: human sumo e1~sumo1-amp tetrahedral intermediate mimic
|
| 10 | c3vh1A_ |
|
 |
100.0 |
24 |
PDB header:metal binding protein
Chain: A: PDB Molecule:ubiquitin-like modifier-activating enzyme atg7;
PDBTitle: crystal structure of saccharomyces cerevisiae atg7 (1-595)
|
| 11 | c1y8qD_ |
|
 |
100.0 |
25 |
PDB header:ligase
Chain: D: PDB Molecule:ubiquitin-like 2 activating enzyme e1b;
PDBTitle: sumo e1 activating enzyme sae1-sae2-mg-atp complex
|
| 12 | c3cmmA_ |
|
 |
100.0 |
27 |
PDB header:ligase/protein binding
Chain: A: PDB Molecule:ubiquitin-activating enzyme e1 1;
PDBTitle: crystal structure of the uba1-ubiquitin complex
|
| 13 | c3gucB_ |
|
 |
100.0 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:ubiquitin-like modifier-activating enzyme 5;
PDBTitle: human ubiquitin-activating enzyme 5 in complex with amppnp
|
| 14 | d1yova1 |
|
 |
100.0 |
19 |
Fold:Activating enzymes of the ubiquitin-like proteins
Superfamily:Activating enzymes of the ubiquitin-like proteins
Family:Ubiquitin activating enzymes (UBA) |
| 15 | c1e5lA_ |
|
 |
98.1 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:saccharopine reductase;
PDBTitle: apo saccharopine reductase from magnaporthe grisea
|
| 16 | d1pjqa1 |
|
 |
98.1 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Siroheme synthase N-terminal domain-like |
| 17 | c2axqA_ |
|
 |
98.1 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:saccharopine dehydrogenase;
PDBTitle: apo histidine-tagged saccharopine dehydrogenase (l-glu2 forming) from saccharomyces cerevisiae
|
| 18 | d1vi2a1 |
|
 |
98.0 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 19 | c3ic5A_ |
|
 |
97.9 |
21 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative saccharopine dehydrogenase;
PDBTitle: n-terminal domain of putative saccharopine dehydrogenase from ruegeria2 pomeroyi.
|
| 20 | c2nloA_ |
|
 |
97.8 |
31 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:shikimate dehydrogenase;
PDBTitle: crystal structure of the quinate dehydrogenase from corynebacterium2 glutamicum
|
| 21 | c1vi2B_ |
|
not modelled |
97.8 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:shikimate 5-dehydrogenase 2;
PDBTitle: crystal structure of shikimate-5-dehydrogenase with nad
|
| 22 | d1pzga1 |
|
not modelled |
97.8 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 23 | c3tozA_ |
|
not modelled |
97.7 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:shikimate dehydrogenase;
PDBTitle: 2.2 angstrom crystal structure of shikimate 5-dehydrogenase from2 listeria monocytogenes in complex with nad.
|
| 24 | c2hjrK_ |
|
not modelled |
97.7 |
24 |
PDB header:oxidoreductase
Chain: K: PDB Molecule:malate dehydrogenase;
PDBTitle: crystal structure of cryptosporidium parvum malate2 dehydrogenase
|
| 25 | c1pjtB_ |
|
not modelled |
97.7 |
21 |
PDB header:transferase/oxidoreductase/lyase
Chain: B: PDB Molecule:siroheme synthase;
PDBTitle: the structure of the ser128ala point-mutant variant of cysg,2 the multifunctional3 methyltransferase/dehydrogenase/ferrochelatase for4 siroheme synthesis
|
| 26 | d1e5qa1 |
|
not modelled |
97.6 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 27 | c2z2vA_ |
|
not modelled |
97.6 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:hypothetical protein ph1688;
PDBTitle: crystal structure of l-lysine dehydrogenase from2 hyperthermophilic archaeon pyrococcus horikoshii
|
| 28 | c1gpjA_ |
|
not modelled |
97.6 |
30 |
PDB header:reductase
Chain: A: PDB Molecule:glutamyl-trna reductase;
PDBTitle: glutamyl-trna reductase from methanopyrus kandleri
|
| 29 | d9ldta1 |
|
not modelled |
97.6 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 30 | d1np3a2 |
|
not modelled |
97.5 |
24 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 31 | c3pgjB_ |
|
not modelled |
97.5 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:shikimate dehydrogenase;
PDBTitle: 2.49 angstrom resolution crystal structure of shikimate 5-2 dehydrogenase (aroe) from vibrio cholerae o1 biovar eltor str. n169613 in complex with shikimate
|
| 32 | c1u4sA_ |
|
not modelled |
97.5 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-lactate dehydrogenase;
PDBTitle: plasmodium falciparum lactate dehydrogenase complexed with 2,6-2 naphthalenedisulphonic acid
|
| 33 | c2fnzA_ |
|
not modelled |
97.5 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:lactate dehydrogenase;
PDBTitle: crystal structure of the lactate dehydrogenase from cryptosporidium2 parvum complexed with cofactor (b-nicotinamide adenine dinucleotide)3 and inhibitor (oxamic acid)
|
| 34 | c2eggA_ |
|
not modelled |
97.5 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:shikimate 5-dehydrogenase;
PDBTitle: crystal structure of shikimate 5-dehydrogenase (aroe) from2 geobacillus kaustophilus
|
| 35 | c1pzfD_ |
|
not modelled |
97.4 |
20 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:lactate dehydrogenase;
PDBTitle: t.gondii ldh1 ternary complex with apad+ and oxalate
|
| 36 | d1uxja1 |
|
not modelled |
97.4 |
25 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 37 | d1t2da1 |
|
not modelled |
97.4 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 38 | d1i0za1 |
|
not modelled |
97.4 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 39 | c1bg6A_ |
|
not modelled |
97.4 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:n-(1-d-carboxylethyl)-l-norvaline dehydrogenase;
PDBTitle: crystal structure of the n-(1-d-carboxylethyl)-l-norvaline2 dehydrogenase from arthrobacter sp. strain 1c
|
| 40 | c3o8qB_ |
|
not modelled |
97.4 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:shikimate 5-dehydrogenase i alpha;
PDBTitle: 1.45 angstrom resolution crystal structure of shikimate 5-2 dehydrogenase (aroe) from vibrio cholerae
|
| 41 | c1ur5C_ |
|
not modelled |
97.4 |
22 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:malate dehydrogenase;
PDBTitle: stabilization of a tetrameric malate dehydrogenase by2 introduction of a disulfide bridge at the dimer/dimer3 interface
|
| 42 | d1gpja2 |
|
not modelled |
97.4 |
30 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 43 | c2g1uA_ |
|
not modelled |
97.4 |
20 |
PDB header:transport protein
Chain: A: PDB Molecule:hypothetical protein tm1088a;
PDBTitle: crystal structure of a putative transport protein (tm1088a) from2 thermotoga maritima at 1.50 a resolution
|
| 44 | c2d0iC_ |
|
not modelled |
97.4 |
18 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:dehydrogenase;
PDBTitle: crystal structure ph0520 protein from pyrococcus horikoshii ot3
|
| 45 | c3u62A_ |
|
not modelled |
97.3 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:shikimate dehydrogenase;
PDBTitle: crystal structure of shikimate dehydrogenase from thermotoga maritima
|
| 46 | c2ew2B_ |
|
not modelled |
97.3 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:2-dehydropantoate 2-reductase, putative;
PDBTitle: crystal structure of the putative 2-dehydropantoate 2-reductase from2 enterococcus faecalis
|
| 47 | c3d1lB_ |
|
not modelled |
97.3 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative nadp oxidoreductase bf3122;
PDBTitle: crystal structure of putative nadp oxidoreductase bf3122 from2 bacteroides fragilis
|
| 48 | c3dfzB_ |
|
not modelled |
97.3 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:precorrin-2 dehydrogenase;
PDBTitle: sirc, precorrin-2 dehydrogenase
|
| 49 | d1lssa_ |
|
not modelled |
97.3 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Potassium channel NAD-binding domain |
| 50 | c3n7uD_ |
|
not modelled |
97.3 |
16 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:formate dehydrogenase;
PDBTitle: nad-dependent formate dehydrogenase from higher-plant arabidopsis2 thaliana in complex with nad and azide
|
| 51 | c3donA_ |
|
not modelled |
97.3 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:shikimate dehydrogenase;
PDBTitle: crystal structure of shikimate dehydrogenase from staphylococcus2 epidermidis
|
| 52 | d1kyqa1 |
|
not modelled |
97.3 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Siroheme synthase N-terminal domain-like |
| 53 | c3eywA_ |
|
not modelled |
97.2 |
24 |
PDB header:transport protein
Chain: A: PDB Molecule:c-terminal domain of glutathione-regulated potassium-efflux
PDBTitle: crystal structure of the c-terminal domain of e. coli kefc in complex2 with keff
|
| 54 | d1pjca1 |
|
not modelled |
97.2 |
32 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 55 | c1np3B_ |
|
not modelled |
97.2 |
26 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:ketol-acid reductoisomerase;
PDBTitle: crystal structure of class i acetohydroxy acid isomeroreductase from2 pseudomonas aeruginosa
|
| 56 | c1z82A_ |
|
not modelled |
97.2 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glycerol-3-phosphate dehydrogenase;
PDBTitle: crystal structure of glycerol-3-phosphate dehydrogenase (tm0378) from2 thermotoga maritima at 2.00 a resolution
|
| 57 | c3gviB_ |
|
not modelled |
97.2 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:malate dehydrogenase;
PDBTitle: crystal structure of lactate/malate dehydrogenase from2 brucella melitensis in complex with adp
|
| 58 | c1m67A_ |
|
not modelled |
97.2 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glycerol-3-phosphate dehydrogenase;
PDBTitle: crystal structure of leishmania mexicana gpdh complexed with inhibitor2 2-bromo-6-hydroxy-purine
|
| 59 | d5ldha1 |
|
not modelled |
97.2 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 60 | c3k96B_ |
|
not modelled |
97.2 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glycerol-3-phosphate dehydrogenase [nad(p)+];
PDBTitle: 2.1 angstrom resolution crystal structure of glycerol-3-phosphate2 dehydrogenase (gpsa) from coxiella burnetii
|
| 61 | d2pgda2 |
|
not modelled |
97.2 |
11 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 62 | d1nvta1 |
|
not modelled |
97.1 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 63 | d1ldna1 |
|
not modelled |
97.1 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 64 | d1i10a1 |
|
not modelled |
97.1 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 65 | c3pwzA_ |
|
not modelled |
97.1 |
32 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:shikimate dehydrogenase 3;
PDBTitle: crystal structure of an ael1 enzyme from pseudomonas putida
|
| 66 | c1ldbA_ |
|
not modelled |
97.1 |
22 |
PDB header:oxidoreductase(choh(d)-nad(a))
Chain: A: PDB Molecule:apo-l-lactate dehydrogenase;
PDBTitle: structure determination and refinement of bacillus2 stearothermophilus lactate dehydrogenase
|
| 67 | c1wpqB_ |
|
not modelled |
97.1 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glycerol-3-phosphate dehydrogenase [nad+],
PDBTitle: ternary complex of glycerol 3-phosphate dehydrogenase 12 with nad and dihydroxyactone
|
| 68 | d1n1ea2 |
|
not modelled |
97.1 |
12 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 69 | c3llvA_ |
|
not modelled |
97.1 |
14 |
PDB header:nad(p) binding protein
Chain: A: PDB Molecule:exopolyphosphatase-related protein;
PDBTitle: the crystal structure of the nad(p)-binding domain of an2 exopolyphosphatase-related protein from archaeoglobus fulgidus to3 1.7a
|
| 70 | d1bg6a2 |
|
not modelled |
97.1 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 71 | d2hmva1 |
|
not modelled |
97.1 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Potassium channel NAD-binding domain |
| 72 | d2ldxa1 |
|
not modelled |
97.0 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 73 | c1wwkA_ |
|
not modelled |
97.0 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:phosphoglycerate dehydrogenase;
PDBTitle: crystal structure of phosphoglycerate dehydrogenase from pyrococcus2 horikoshii ot3
|
| 74 | c3nzoB_ |
|
not modelled |
97.0 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:udp-n-acetylglucosamine 4,6-dehydratase;
PDBTitle: udp-n-acetylglucosamine 4,6-dehydratase from vibrio fischeri.
|
| 75 | c2d4aC_ |
|
not modelled |
97.0 |
20 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:malate dehydrogenase;
PDBTitle: structure of the malate dehydrogenase from aeropyrum pernix
|
| 76 | c1hyhA_ |
|
not modelled |
97.0 |
27 |
PDB header:oxidoreductase (choh(d)-nad+(a))
Chain: A: PDB Molecule:l-2-hydroxyisocaproate dehydrogenase;
PDBTitle: crystal structure of l-2-hydroxyisocaproate dehydrogenase from2 lactobacillus confusus at 2.2 angstroms resolution-an example of3 strong asymmetry between subunits
|
| 77 | c1gv1D_ |
|
not modelled |
97.0 |
23 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:malate dehydrogenase;
PDBTitle: structural basis for thermophilic protein stability:2 structures of thermophilic and mesophilic malate3 dehydrogenases
|
| 78 | c8ldhA_ |
|
not modelled |
97.0 |
14 |
PDB header:oxidoreductase(choh(d)-nad(a))
Chain: A: PDB Molecule:m4 apo-lactate dehydrogenase;
PDBTitle: refined crystal structure of dogfish m4 apo-lactate2 dehydrogenase
|
| 79 | c3cumA_ |
|
not modelled |
97.0 |
31 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable 3-hydroxyisobutyrate dehydrogenase;
PDBTitle: crystal structure of a possible 3-hydroxyisobutyrate dehydrogenase2 from pseudomonas aeruginosa pao1
|
| 80 | c1kyqC_ |
|
not modelled |
97.0 |
16 |
PDB header:oxidoreductase, lyase
Chain: C: PDB Molecule:siroheme biosynthesis protein met8;
PDBTitle: met8p: a bifunctional nad-dependent dehydrogenase and2 ferrochelatase involved in siroheme synthesis.
|
| 81 | c2dfdD_ |
|
not modelled |
97.0 |
34 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:malate dehydrogenase;
PDBTitle: crystal structure of human malate dehydrogenase type 2
|
| 82 | c3pqeD_ |
|
not modelled |
97.0 |
24 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:l-lactate dehydrogenase;
PDBTitle: crystal structure of l-lactate dehydrogenase from bacillus subtilis2 with h171c mutation
|
| 83 | c2ldxA_ |
|
not modelled |
96.9 |
22 |
PDB header:oxidoreductase(choh(d)-nad(a))
Chain: A: PDB Molecule:apo-lactate dehydrogenase;
PDBTitle: characterization of the antigenic sites on the refined 3-2 angstroms resolution structure of mouse testicular lactate3 dehydrogenase c4
|
| 84 | c3k6jA_ |
|
not modelled |
96.9 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein f01g10.3, confirmed by transcript evidence;
PDBTitle: crystal structure of the dehydrogenase part of multifuctional enzyme 12 from c.elegans
|
| 85 | c2ph5A_ |
|
not modelled |
96.9 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:homospermidine synthase;
PDBTitle: crystal structure of the homospermidine synthase hss from legionella2 pneumophila in complex with nad, northeast structural genomics target3 lgr54
|
| 86 | c1pgqA_ |
|
not modelled |
96.9 |
9 |
PDB header:oxidoreductase (choh(d)-nadp+(a))
Chain: A: PDB Molecule:6-phosphogluconate dehydrogenase;
PDBTitle: crystallographic study of coenzyme, coenzyme analogue and substrate2 binding in 6-phosphogluconate dehydrogenase: implications for nadp3 specificity and the enzyme mechanism
|
| 87 | c1txgA_ |
|
not modelled |
96.9 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glycerol-3-phosphate dehydrogenase [nad(p)+];
PDBTitle: structure of glycerol-3-phosphate dehydrogenase from archaeoglobus2 fulgidus
|
| 88 | c2nacA_ |
|
not modelled |
96.9 |
14 |
PDB header:oxidoreductase(aldehyde(d),nad+(a))
Chain: A: PDB Molecule:nad-dependent formate dehydrogenase;
PDBTitle: high resolution structures of holo and apo formate dehydrogenase
|
| 89 | d1llda1 |
|
not modelled |
96.9 |
25 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 90 | c3hwrA_ |
|
not modelled |
96.9 |
28 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2-dehydropantoate 2-reductase;
PDBTitle: crystal structure of pane/apba family ketopantoate reductase2 (yp_299159.1) from ralstonia eutropha jmp134 at 2.15 a resolution
|
| 91 | d1obba1 |
|
not modelled |
96.9 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 92 | c2ofpB_ |
|
not modelled |
96.9 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:ketopantoate reductase;
PDBTitle: crystal structure of escherichia coli ketopantoate2 reductase in a ternary complex with nadp+ and pantoate
|
| 93 | d1hyha1 |
|
not modelled |
96.9 |
32 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 94 | c3evtA_ |
|
not modelled |
96.9 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:phosphoglycerate dehydrogenase;
PDBTitle: crystal structure of phosphoglycerate dehydrogenase from2 lactobacillus plantarum
|
| 95 | c3djeA_ |
|
not modelled |
96.9 |
38 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:fructosyl amine: oxygen oxidoreductase;
PDBTitle: crystal structure of the deglycating enzyme fructosamine2 oxidase from aspergillus fumigatus (amadoriase ii) in3 complex with fsa
|
| 96 | c1a5zA_ |
|
not modelled |
96.9 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-lactate dehydrogenase;
PDBTitle: lactate dehydrogenase from thermotoga maritima (tmldh)
|
| 97 | d1s6ya1 |
|
not modelled |
96.9 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 98 | c1ojuA_ |
|
not modelled |
96.9 |
9 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:malate dehydrogenase;
PDBTitle: 2.8 a resolution structure of malate dehydrogenase from2 archaeoglobus fulgidus in complex with etheno-nad.
|
| 99 | c2gcgB_ |
|
not modelled |
96.9 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glyoxylate reductase/hydroxypyruvate reductase;
PDBTitle: ternary crystal structure of human glyoxylate2 reductase/hydroxypyruvate reductase
|
| 100 | d1gtea4 |
|
not modelled |
96.8 |
8 |
Fold:Nucleotide-binding domain
Superfamily:Nucleotide-binding domain
Family:N-terminal domain of adrenodoxin reductase-like |
| 101 | d1u8xx1 |
|
not modelled |
96.8 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 102 | c3d0oA_ |
|
not modelled |
96.8 |
28 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-lactate dehydrogenase 1;
PDBTitle: crystal structure of lactate dehydrogenase from2 staphylococcus aureus
|
| 103 | c1pj6A_ |
|
not modelled |
96.8 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:n,n-dimethylglycine oxidase;
PDBTitle: crystal structure of dimethylglycine oxidase of arthrobacter2 globiformis in complex with folic acid
|
| 104 | c2dbqA_ |
|
not modelled |
96.8 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glyoxylate reductase;
PDBTitle: crystal structure of glyoxylate reductase (ph0597) from pyrococcus2 horikoshii ot3, complexed with nadp (i41)
|
| 105 | c1nvtA_ |
|
not modelled |
96.8 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:shikimate 5'-dehydrogenase;
PDBTitle: crystal structure of shikimate dehydrogenase (aroe or2 mj1084) in complex with nadp+
|
| 106 | c2e37B_ |
|
not modelled |
96.8 |
27 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:l-lactate dehydrogenase;
PDBTitle: structure of tt0471 protein from thermus thermophilus
|
| 107 | c2v65A_ |
|
not modelled |
96.8 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-lactate dehydrogenase a chain;
PDBTitle: apo ldh from the psychrophile c. gunnari
|
| 108 | d1txga2 |
|
not modelled |
96.8 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 109 | c1mldA_ |
|
not modelled |
96.8 |
33 |
PDB header:oxidoreductase(nad(a)-choh(d))
Chain: A: PDB Molecule:malate dehydrogenase;
PDBTitle: refined structure of mitochondrial malate dehydrogenase2 from porcine heart and the consensus structure for3 dicarboxylic acid oxidoreductases
|
| 110 | d1ojua1 |
|
not modelled |
96.8 |
12 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 111 | c3fi9B_ |
|
not modelled |
96.8 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:malate dehydrogenase;
PDBTitle: crystal structure of malate dehydrogenase from porphyromonas2 gingivalis
|
| 112 | c2g76A_ |
|
not modelled |
96.8 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-3-phosphoglycerate dehydrogenase;
PDBTitle: crystal structure of human 3-phosphoglycerate dehydrogenase
|
| 113 | c3qhaB_ |
|
not modelled |
96.7 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of a putative oxidoreductase from mycobacterium2 avium 104
|
| 114 | c2cukC_ |
|
not modelled |
96.7 |
21 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:glycerate dehydrogenase/glyoxylate reductase;
PDBTitle: crystal structure of tt0316 protein from thermus thermophilus hb8
|
| 115 | d1ldma1 |
|
not modelled |
96.7 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 116 | c1lldA_ |
|
not modelled |
96.7 |
18 |
PDB header:oxidoreductase(choh (d)-nad (a))
Chain: A: PDB Molecule:l-lactate dehydrogenase;
PDBTitle: molecular basis of allosteric activation of bacterial l-lactate2 dehydrogenase
|
| 117 | c1ks9A_ |
|
not modelled |
96.7 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2-dehydropantoate 2-reductase;
PDBTitle: ketopantoate reductase from escherichia coli
|
| 118 | d2jfga1 |
|
not modelled |
96.7 |
20 |
Fold:MurCD N-terminal domain
Superfamily:MurCD N-terminal domain
Family:MurCD N-terminal domain |
| 119 | c1pgjA_ |
|
not modelled |
96.7 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:6-phosphogluconate dehydrogenase;
PDBTitle: x-ray structure of 6-phosphogluconate dehydrogenase from the protozoan2 parasite t. brucei
|
| 120 | c2iz1C_ |
|
not modelled |
96.7 |
14 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:6-phosphogluconate dehydrogenase, decarboxylating;
PDBTitle: 6pdh complexed with pex inhibitor synchrotron data
|

