| 1 | d1u7za_
|
|
 |
100.0 |
99 |
Fold:Ribokinase-like
Superfamily:CoaB-like
Family:CoaB-like |
| 2 | c2gk4A_
|
|
 |
100.0 |
27 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:conserved hypothetical protein;
PDBTitle: the crystal structure of the dna/pantothenate metabolism flavoprotein2 from streptococcus pneumoniae
|
| 3 | d1p9oa_
|
|
 |
100.0 |
21 |
Fold:Ribokinase-like
Superfamily:CoaB-like
Family:CoaB-like |
| 4 | d1g5qa_
|
|
 |
100.0 |
19 |
Fold:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
Superfamily:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
Family:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD |
| 5 | d1p3y1_
|
|
 |
100.0 |
26 |
Fold:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
Superfamily:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
Family:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD |
| 6 | c3qjgD_
|
|
 |
100.0 |
20 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:epidermin biosynthesis protein epid;
PDBTitle: epidermin biosynthesis protein epid from staphylococcus aureus
|
| 7 | c1mvlA_
|
|
 |
100.0 |
28 |
PDB header:lyase
Chain: A: PDB Molecule:ppc decarboxylase athal3a;
PDBTitle: ppc decarboxylase mutant c175s
|
| 8 | d1mvla_
|
|
 |
100.0 |
28 |
Fold:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
Superfamily:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
Family:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD |
| 9 | c3mcuF_
|
|
 |
100.0 |
19 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:dipicolinate synthase, b chain;
PDBTitle: crystal structure of the dipicolinate synthase chain b from2 bacillus cereus. northeast structural genomics consortium3 target bcr215.
|
| 10 | c3lqkA_
|
|
 |
100.0 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dipicolinate synthase subunit b;
PDBTitle: crystal structure of dipicolinate synthase subunit b from bacillus2 halodurans c
|
| 11 | d1qzua_
|
|
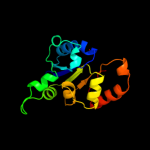 |
100.0 |
29 |
Fold:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
Superfamily:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
Family:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD |
| 12 | d1sbza_
|
|
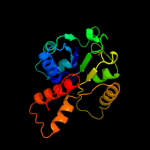 |
100.0 |
19 |
Fold:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
Superfamily:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
Family:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD |
| 13 | c2ejbA_
|
|
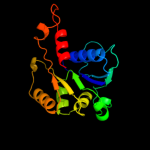 |
100.0 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:probable aromatic acid decarboxylase;
PDBTitle: crystal structure of phenylacrylic acid decarboxylase from2 aquifex aeolicus
|
| 14 | c1qzuB_
|
|
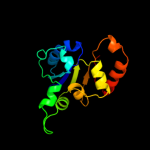 |
100.0 |
29 |
PDB header:lyase
Chain: B: PDB Molecule:hypothetical protein mds018;
PDBTitle: crystal structure of human phosphopantothenoylcysteine decarboxylase
|
| 15 | c3zquA_
|
|
 |
100.0 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:probable aromatic acid decarboxylase;
PDBTitle: structure of a probable aromatic acid decarboxylase
|
| 16 | c2et6A_
|
|
 |
98.2 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:(3r)-hydroxyacyl-coa dehydrogenase;
PDBTitle: (3r)-hydroxyacyl-coa dehydrogenase domain of candida tropicalis2 peroxisomal multifunctional enzyme type 2
|
| 17 | d1rkxa_
|
|
 |
97.7 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 18 | c2q1uA_
|
|
 |
97.6 |
19 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:putative nucleotide sugar epimerase/ dehydratase;
PDBTitle: crystal structure of the bordetella bronchiseptica enzyme wbmf in2 complex with nad+ and udp
|
| 19 | c1z7eC_
|
|
 |
97.6 |
17 |
PDB header:hydrolase
Chain: C: PDB Molecule:protein arna;
PDBTitle: crystal structure of full length arna
|
| 20 | d1wvga1
|
|
 |
97.6 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 21 | c3nzoB_ |
|
not modelled |
97.5 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:udp-n-acetylglucosamine 4,6-dehydratase;
PDBTitle: udp-n-acetylglucosamine 4,6-dehydratase from vibrio fischeri.
|
| 22 | c3d4oA_ |
|
not modelled |
97.4 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dipicolinate synthase subunit a;
PDBTitle: crystal structure of dipicolinate synthase subunit a (np_243269.1)2 from bacillus halodurans at 2.10 a resolution
|
| 23 | c3qvoA_ |
|
not modelled |
97.3 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:nmra family protein;
PDBTitle: structure of a rossmann-fold nad(p)-binding family protein from2 shigella flexneri.
|
| 24 | c3grkE_ |
|
not modelled |
97.3 |
11 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:enoyl-(acyl-carrier-protein) reductase (nadh);
PDBTitle: crystal structure of short chain dehydrogenase reductase2 sdr glucose-ribitol dehydrogenase from brucella melitensis
|
| 25 | c3lu1C_ |
|
not modelled |
97.3 |
11 |
PDB header:isomerase
Chain: C: PDB Molecule:wbgu;
PDBTitle: crystal structure analysis of wbgu: a udp-galnac 4-epimerase
|
| 26 | d1db3a_ |
|
not modelled |
97.3 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 27 | c2z1mC_ |
|
not modelled |
97.3 |
20 |
PDB header:lyase
Chain: C: PDB Molecule:gdp-d-mannose dehydratase;
PDBTitle: crystal structure of gdp-d-mannose dehydratase from aquifex aeolicus2 vf5
|
| 28 | c2ptgA_ |
|
not modelled |
97.2 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:enoyl-acyl carrier reductase;
PDBTitle: crystal structure of eimeria tenella enoyl reductase
|
| 29 | d1luaa1 |
|
not modelled |
97.2 |
24 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 30 | c3ctmH_ |
|
not modelled |
97.2 |
20 |
PDB header:oxidoreductase
Chain: H: PDB Molecule:carbonyl reductase;
PDBTitle: crystal structure of a carbonyl reductase from candida2 parapsilosis with anti-prelog stereo-specificity
|
| 31 | c2fwmX_ |
|
not modelled |
97.1 |
27 |
PDB header:oxidoreductase
Chain: X: PDB Molecule:2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase;
PDBTitle: crystal structure of e. coli enta, a 2,3-dihydrodihydroxy benzoate2 dehydrogenase
|
| 32 | c1zbqB_ |
|
not modelled |
97.1 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:17-beta-hydroxysteroid dehydrogenase 4;
PDBTitle: crystal structure of human 17-beta-hydroxysteroid dehydrogenase type 42 in complex with nad
|
| 33 | d1zbqa1 |
|
not modelled |
97.1 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 34 | c1luaA_ |
|
not modelled |
97.1 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:methylene tetrahydromethanopterin dehydrogenase;
PDBTitle: structure of methylene-tetrahydromethanopterin dehydrogenase from2 methylobacterium extorquens am1 complexed with nadp
|
| 35 | c3un1D_ |
|
not modelled |
97.1 |
17 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:probable oxidoreductase;
PDBTitle: crystal structure of an oxidoreductase from sinorhizobium meliloti2 1021
|
| 36 | d1e6ua_ |
|
not modelled |
97.1 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 37 | c2dteB_ |
|
not modelled |
97.1 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glucose 1-dehydrogenase related protein;
PDBTitle: structure of thermoplasma acidophilum aldohexose dehydrogenase (aldt)2 in complex with nadh
|
| 38 | c1z45A_ |
|
not modelled |
97.0 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:gal10 bifunctional protein;
PDBTitle: crystal structure of the gal10 fusion protein galactose2 mutarotase/udp-galactose 4-epimerase from saccharomyces3 cerevisiae complexed with nad, udp-glucose, and galactose
|
| 39 | c3i1jB_ |
|
not modelled |
97.0 |
35 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase, short chain
PDBTitle: structure of a putative short chain dehydrogenase from2 pseudomonas syringae
|
| 40 | d1ulua_ |
|
not modelled |
97.0 |
24 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 41 | d1hdoa_ |
|
not modelled |
97.0 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 42 | c3tpcG_ |
|
not modelled |
97.0 |
21 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:short chain alcohol dehydrogenase-related dehydrogenase;
PDBTitle: crystal structure of a hypothtical protein sma1452 from sinorhizobium2 meliloti 1021
|
| 43 | c2qioA_ |
|
not modelled |
97.0 |
14 |
PDB header:unknown function
Chain: A: PDB Molecule:enoyl-(acyl-carrier-protein) reductase;
PDBTitle: x-ray structure of enoyl-acyl carrier protein reductase from bacillus2 anthracis with triclosan
|
| 44 | d1zk4a1 |
|
not modelled |
97.0 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 45 | d1hdca_ |
|
not modelled |
97.0 |
37 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 46 | d1yxma1 |
|
not modelled |
97.0 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 47 | c2rirA_ |
|
not modelled |
97.0 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dipicolinate synthase, a chain;
PDBTitle: crystal structure of dipicolinate synthase, a chain, from bacillus2 subtilis
|
| 48 | d1qsga_ |
|
not modelled |
97.0 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 49 | c3ek2D_ |
|
not modelled |
97.0 |
17 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:enoyl-(acyl-carrier-protein) reductase (nadh);
PDBTitle: crystal structure of eonyl-(acyl carrier protein) reductase2 from burkholderia pseudomallei 1719b
|
| 50 | d2pd4a1 |
|
not modelled |
97.0 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 51 | d1n7ha_ |
|
not modelled |
97.0 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 52 | c3t7cC_ |
|
not modelled |
97.0 |
19 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:carveol dehydrogenase;
PDBTitle: crystal structure of carveol dehydrogenase from mycobacterium avium2 bound to nad
|
| 53 | c2x4gA_ |
|
not modelled |
97.0 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:nucleoside-diphosphate-sugar epimerase;
PDBTitle: crystal structure of pa4631, a nucleoside-diphosphate-sugar2 epimerase from pseudomonas aeruginosa
|
| 54 | c3uveC_ |
|
not modelled |
97.0 |
16 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:carveol dehydrogenase ((+)-trans-carveol dehydrogenase);
PDBTitle: crystal structure of carveol dehydrogenase ((+)-trans-carveol2 dehydrogenase) from mycobacterium avium
|
| 55 | d1gz6a_ |
|
not modelled |
96.9 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 56 | c3gvcB_ |
|
not modelled |
96.9 |
25 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable short-chain type
PDBTitle: crystal structure of probable short-chain dehydrogenase-2 reductase from mycobacterium tuberculosis
|
| 57 | d1vl8a_ |
|
not modelled |
96.9 |
38 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 58 | c3lf2B_ |
|
not modelled |
96.9 |
26 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:short chain oxidoreductase q9hya2;
PDBTitle: nadph bound structure of the short chain oxidoreductase q9hya2 from2 pseudomonas aeruginosa pao1 containing an atypical catalytic center
|
| 59 | c2gn9B_ |
|
not modelled |
96.9 |
23 |
PDB header:lyase
Chain: B: PDB Molecule:udp-glcnac c6 dehydratase;
PDBTitle: crystal structure of udp-glcnac inverting 4,6-dehydratase in complex2 with nadp and udp-glc
|
| 60 | c2jahB_ |
|
not modelled |
96.9 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:clavulanic acid dehydrogenase;
PDBTitle: biochemical and structural analysis of the clavulanic acid2 dehydeogenase (cad) from streptomyces clavuligerus
|
| 61 | c1n7gB_ |
|
not modelled |
96.9 |
19 |
PDB header:lyase
Chain: B: PDB Molecule:gdp-d-mannose-4,6-dehydratase;
PDBTitle: crystal structure of the gdp-mannose 4,6-dehydratase2 ternary complex with nadph and gdp-rhamnose.
|
| 62 | c3rfxB_ |
|
not modelled |
96.9 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:uronate dehydrogenase;
PDBTitle: crystal structure of uronate dehydrogenase from agrobacterium2 tumefaciens, y136a mutant complexed with nad
|
| 63 | c3gdfA_ |
|
not modelled |
96.9 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable nadp-dependent mannitol dehydrogenase;
PDBTitle: crystal structure of the nadp-dependent mannitol dehydrogenase from2 cladosporium herbarum.
|
| 64 | d1i24a_ |
|
not modelled |
96.9 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 65 | d2o23a1 |
|
not modelled |
96.9 |
38 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 66 | c3ppiA_ |
|
not modelled |
96.8 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-hydroxyacyl-coa dehydrogenase type-2;
PDBTitle: crystal structure of 3-hydroxyacyl-coa dehydrogenase type-2 from2 mycobacterium avium
|
| 67 | c3ioyB_ |
|
not modelled |
96.8 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:short-chain dehydrogenase/reductase sdr;
PDBTitle: structure of putative short-chain dehydrogenase (saro_0793)2 from novosphingobium aromaticivorans
|
| 68 | c3svtA_ |
|
not modelled |
96.8 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:short-chain type dehydrogenase/reductase;
PDBTitle: structure of a short-chain type dehydrogenase/reductase from2 mycobacterium ulcerans
|
| 69 | d2h7ma1 |
|
not modelled |
96.8 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 70 | c2uvdE_ |
|
not modelled |
96.8 |
23 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:3-oxoacyl-(acyl-carrier-protein) reductase;
PDBTitle: the crystal structure of a 3-oxoacyl-(acyl carrier protein)2 reductase from bacillus anthracis (ba3989)
|
| 71 | d1cyda_ |
|
not modelled |
96.8 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 72 | c2pk3B_ |
|
not modelled |
96.8 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:gdp-6-deoxy-d-lyxo-4-hexulose reductase;
PDBTitle: crystal structure of a gdp-4-keto-6-deoxy-d-mannose reductase
|
| 73 | c2ydyA_ |
|
not modelled |
96.8 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:methionine adenosyltransferase 2 subunit beta;
PDBTitle: crystal structure of human s-adenosylmethionine synthetase2 2, beta subunit in orthorhombic crystal form
|
| 74 | d1uzma1 |
|
not modelled |
96.8 |
24 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 75 | d1ae1a_ |
|
not modelled |
96.8 |
27 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 76 | c3pgxB_ |
|
not modelled |
96.8 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:carveol dehydrogenase;
PDBTitle: crystal structure of a putative carveol dehydrogenase from2 mycobacterium paratuberculosis bound to nicotinamide adenine3 dinucleotide
|
| 77 | c3oh8A_ |
|
not modelled |
96.8 |
19 |
PDB header:isomerase
Chain: A: PDB Molecule:nucleoside-diphosphate sugar epimerase (sula family);
PDBTitle: crystal structure of the nucleoside-diphosphate sugar epimerase from2 corynebacterium glutamicum. northeast structural genomics consortium3 target cgr91
|
| 78 | c3ksuA_ |
|
not modelled |
96.8 |
29 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-oxoacyl-acyl carrier protein reductase;
PDBTitle: crystal structure of short-chain dehydrogenase from2 oenococcus oeni psu-1
|
| 79 | c3rd5A_ |
|
not modelled |
96.8 |
32 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:mypaa.01249.c;
PDBTitle: crystal structure of a putative uncharacterized protein from2 mycobacterium paratuberculosis
|
| 80 | d1h5qa_ |
|
not modelled |
96.8 |
24 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 81 | c3gr6A_ |
|
not modelled |
96.7 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:enoyl-[acyl-carrier-protein] reductase [nadh];
PDBTitle: crystal structure of the staphylococcus aureus enoyl-acyl2 carrier protein reductase (fabi) in complex with nadp and3 triclosan
|
| 82 | c3e8xA_ |
|
not modelled |
96.7 |
10 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative nad-dependent epimerase/dehydratase;
PDBTitle: putative nad-dependent epimerase/dehydratase from bacillus halodurans.
|
| 83 | d1o5ia_ |
|
not modelled |
96.7 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 84 | c2cfcB_ |
|
not modelled |
96.7 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:2-(r)-hydroxypropyl-com dehydrogenase;
PDBTitle: structural basis for stereo selectivity in the (r)- and2 (s)-hydroxypropylethane thiosulfonate dehydrogenases
|
| 85 | d1bxka_ |
|
not modelled |
96.7 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 86 | d2c5aa1 |
|
not modelled |
96.7 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 87 | c3f9iB_ |
|
not modelled |
96.7 |
35 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:3-oxoacyl-[acyl-carrier-protein] reductase;
PDBTitle: crystal structure of 3-ketoacyl-(acyl-carrier-protein) reductase2 rickettsia prowazekii
|
| 88 | d1xg5a_ |
|
not modelled |
96.7 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 89 | d1ek6a_ |
|
not modelled |
96.7 |
12 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 90 | c3ftpD_ |
|
not modelled |
96.7 |
36 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:3-oxoacyl-[acyl-carrier protein] reductase;
PDBTitle: crystal structure of 3-ketoacyl-(acyl-carrier-protein)2 reductase from burkholderia pseudomallei at 2.05 a3 resolution
|
| 91 | c3k31B_ |
|
not modelled |
96.7 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:enoyl-(acyl-carrier-protein) reductase;
PDBTitle: crystal structure of eonyl-(acyl-carrier-protein) reductase from2 anaplasma phagocytophilum in complex with nad at 1.9a resolution
|
| 92 | c2q1wC_ |
|
not modelled |
96.7 |
21 |
PDB header:sugar binding protein
Chain: C: PDB Molecule:putative nucleotide sugar epimerase/ dehydratase;
PDBTitle: crystal structure of the bordetella bronchiseptica enzyme wbmh in2 complex with nad+
|
| 93 | c3rkrC_ |
|
not modelled |
96.7 |
33 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:short chain oxidoreductase;
PDBTitle: crystal structure of a metagenomic short-chain oxidoreductase (sdr) in2 complex with nadp
|
| 94 | c2ntnB_ |
|
not modelled |
96.7 |
26 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:3-oxoacyl-[acyl-carrier-protein] reductase;
PDBTitle: crystal structure of maba-c60v/g139a/s144l
|
| 95 | c3pk0B_ |
|
not modelled |
96.7 |
23 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:short-chain dehydrogenase/reductase sdr;
PDBTitle: crystal structure of short-chain dehydrogenase/reductase sdr from2 mycobacterium smegmatis
|
| 96 | c2o2sA_ |
|
not modelled |
96.7 |
38 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:enoyl-acyl carrier reductase;
PDBTitle: the structure of t. gondii enoyl acyl carrier protein reductase in2 complex with nad and triclosan
|
| 97 | d1a4ia1 |
|
not modelled |
96.7 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 98 | c3afnC_ |
|
not modelled |
96.7 |
43 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:carbonyl reductase;
PDBTitle: crystal structure of aldose reductase a1-r complexed with nadp
|
| 99 | c2zatC_ |
|
not modelled |
96.7 |
37 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:dehydrogenase/reductase sdr family member 4;
PDBTitle: crystal structure of a mammalian reductase
|
| 100 | c3toxG_ |
|
not modelled |
96.6 |
18 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:short chain dehydrogenase;
PDBTitle: crystal structure of a short chain dehydrogenase in complex with2 nad(p) from sinorhizobium meliloti 1021
|
| 101 | c2jjyD_ |
|
not modelled |
96.6 |
16 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:enoyl-[acyl-carrier-protein] reductase;
PDBTitle: crystal structure of francisella tularensis enoyl reductase2 (ftfabi) with bound nad
|
| 102 | d1q7ba_ |
|
not modelled |
96.6 |
30 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 103 | c3r3sD_ |
|
not modelled |
96.6 |
42 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:oxidoreductase;
PDBTitle: structure of the ygha oxidoreductase from salmonella enterica
|
| 104 | c3emkA_ |
|
not modelled |
96.6 |
35 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glucose/ribitol dehydrogenase;
PDBTitle: 2.5a crystal structure of glucose/ribitol dehydrogenase2 from brucella melitensis
|
| 105 | c3o26A_ |
|
not modelled |
96.6 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:salutaridine reductase;
PDBTitle: the structure of salutaridine reductase from papaver somniferum.
|
| 106 | d1kewa_ |
|
not modelled |
96.6 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 107 | c3itdA_ |
|
not modelled |
96.6 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:17beta-hydroxysteroid dehydrogenase;
PDBTitle: crystal structure of an inactive 17beta-hydroxysteroid dehydrogenase2 (y167f mutated form) from fungus cochliobolus lunatus
|
| 108 | d1ja9a_ |
|
not modelled |
96.6 |
31 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 109 | d1pr9a_ |
|
not modelled |
96.6 |
27 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 110 | c3pxxE_ |
|
not modelled |
96.6 |
18 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:carveol dehydrogenase;
PDBTitle: crystal structure of carveol dehydrogenase from mycobacterium avium2 bound to nicotinamide adenine dinucleotide
|
| 111 | d1qyda_ |
|
not modelled |
96.6 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 112 | c2hq1A_ |
|
not modelled |
96.6 |
41 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glucose/ribitol dehydrogenase;
PDBTitle: crystal structure of orf 1438 a putative glucose/ribitol2 dehydrogenase from clostridium thermocellum
|
| 113 | c3ijrF_ |
|
not modelled |
96.6 |
20 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:oxidoreductase, short chain dehydrogenase/reductase family;
PDBTitle: 2.05 angstrom resolution crystal structure of a short chain2 dehydrogenase from bacillus anthracis str. 'ames ancestor' in complex3 with nad+
|
| 114 | d1ooea_ |
|
not modelled |
96.6 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 115 | c3uxyC_ |
|
not modelled |
96.6 |
24 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:short-chain dehydrogenase/reductase sdr;
PDBTitle: the crystal structure of short chain dehydrogenase from rhodobacter2 sphaeroides
|
| 116 | c3dhnA_ |
|
not modelled |
96.6 |
17 |
PDB header:isomerase, lyase
Chain: A: PDB Molecule:nad-dependent epimerase/dehydratase;
PDBTitle: crystal structure of the putative epimerase q89z24_bactn2 from bacteroides thetaiotaomicron. northeast structural3 genomics consortium target btr310.
|
| 117 | d1yb1a_ |
|
not modelled |
96.5 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 118 | c3s55F_ |
|
not modelled |
96.5 |
31 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:putative short-chain dehydrogenase/reductase;
PDBTitle: crystal structure of a putative short-chain dehydrogenase/reductase2 from mycobacterium abscessus bound to nad
|
| 119 | c3slgB_ |
|
not modelled |
96.5 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:pbgp3 protein;
PDBTitle: crystal structure of pbgp3 protein from burkholderia pseudomallei
|
| 120 | c3c1oA_ |
|
not modelled |
96.5 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:eugenol synthase;
PDBTitle: the multiple phenylpropene synthases in both clarkia2 breweri and petunia hybrida represent two distinct lineages
|