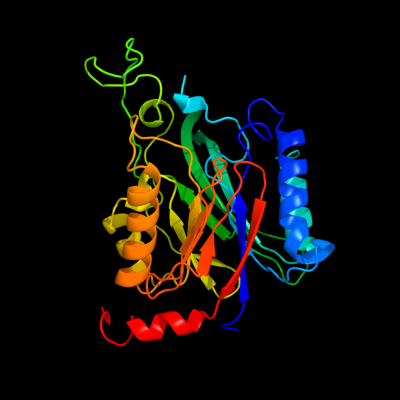
1 d1uf5a_
100.0
18
Fold: Carbon-nitrogen hydrolaseSuperfamily: Carbon-nitrogen hydrolaseFamily: Carbamilase2 c2plqA_
100.0
15
PDB header: hydrolaseChain: A: PDB Molecule: aliphatic amidase;PDBTitle: crystal structure of the amidase from geobacillus pallidus rapc8
3 c2vhiG_
100.0
17
PDB header: hydrolaseChain: G: PDB Molecule: cg3027-pa;PDBTitle: crystal structure of a pyrimidine degrading enzyme from2 drosophila melanogaster
4 c1emsB_
100.0
17
PDB header: antitumor proteinChain: B: PDB Molecule: nit-fragile histidine triad fusion protein;PDBTitle: crystal structure of the c. elegans nitfhit protein
5 c2e2kC_
100.0
18
PDB header: hydrolaseChain: C: PDB Molecule: formamidase;PDBTitle: helicobacter pylori formamidase amif contains a fine-tuned cysteine-2 glutamate-lysine catalytic triad
6 c2w1vA_
100.0
15
PDB header: hydrolaseChain: A: PDB Molecule: nitrilase homolog 2;PDBTitle: crystal structure of mouse nitrilase-2 at 1.4a resolution
7 d1emsa2
100.0
17
Fold: Carbon-nitrogen hydrolaseSuperfamily: Carbon-nitrogen hydrolaseFamily: Nitrilase8 c3hkxA_
100.0
22
PDB header: hydrolaseChain: A: PDB Molecule: amidase;PDBTitle: crystal structure analysis of an amidase from nesterenkonia sp.
9 d1f89a_
100.0
22
Fold: Carbon-nitrogen hydrolaseSuperfamily: Carbon-nitrogen hydrolaseFamily: Nitrilase10 c2e11B_
100.0
22
PDB header: hydrolaseChain: B: PDB Molecule: hydrolase;PDBTitle: the crystal structure of xc1258 from xanthomonas campestris: a cn-2 hydrolase superfamily protein with an arsenic adduct in the active3 site
11 d1j31a_
100.0
17
Fold: Carbon-nitrogen hydrolaseSuperfamily: Carbon-nitrogen hydrolaseFamily: Carbamilase12 c3n05B_
100.0
20
PDB header: ligaseChain: B: PDB Molecule: nh(3)-dependent nad(+) synthetase;PDBTitle: crystal structure of nh3-dependent nad+ synthetase from streptomyces2 avermitilis
13 c3dlaD_
100.0
17
PDB header: ligaseChain: D: PDB Molecule: glutamine-dependent nad(+) synthetase;PDBTitle: x-ray crystal structure of glutamine-dependent nad+ synthetase from2 mycobacterium tuberculosis bound to naad+ and don
14 c3ilvA_
100.0
14
PDB header: ligaseChain: A: PDB Molecule: glutamine-dependent nad(+) synthetase;PDBTitle: crystal structure of a glutamine-dependent nad(+) synthetase2 from cytophaga hutchinsonii
15 d1sgva1
74.4
19
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: PUA domain16 d4pgaa_
58.7
13
Fold: Glutaminase/AsparaginaseSuperfamily: Glutaminase/AsparaginaseFamily: Glutaminase/Asparaginase17 d1zq1a2
47.0
12
Fold: Glutaminase/AsparaginaseSuperfamily: Glutaminase/AsparaginaseFamily: Glutaminase/Asparaginase18 c2wltA_
45.4
13
PDB header: hydrolaseChain: A: PDB Molecule: l-asparaginase;PDBTitle: the crystal structure of helicobacter pylori l-asparaginase2 at 1.4 a resolution
19 d2d6fa2
43.1
10
Fold: Glutaminase/AsparaginaseSuperfamily: Glutaminase/AsparaginaseFamily: Glutaminase/Asparaginase20 c1xf1A_
42.3
12
PDB header: hydrolaseChain: A: PDB Molecule: c5a peptidase;PDBTitle: structure of c5a peptidase- a key virulence factor from2 streptococcus
21 d1o7ja_
not modelled
40.0
16
Fold: Glutaminase/AsparaginaseSuperfamily: Glutaminase/AsparaginaseFamily: Glutaminase/Asparaginase22 c1zq1B_
not modelled
39.9
13
PDB header: lyaseChain: B: PDB Molecule: glutamyl-trna(gln) amidotransferase subunit d;PDBTitle: structure of gatde trna-dependent amidotransferase from2 pyrococcus abyssi
23 d3dhwc1
not modelled
38.7
12
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like24 d1nnsa_
not modelled
36.4
11
Fold: Glutaminase/AsparaginaseSuperfamily: Glutaminase/AsparaginaseFamily: Glutaminase/Asparaginase25 d2ocda1
not modelled
32.3
16
Fold: Glutaminase/AsparaginaseSuperfamily: Glutaminase/AsparaginaseFamily: Glutaminase/Asparaginase26 c2d6fA_
not modelled
29.9
10
PDB header: ligase/rnaChain: A: PDB Molecule: glutamyl-trna(gln) amidotransferase subunit d;PDBTitle: crystal structure of glu-trna(gln) amidotransferase in the2 complex with trna(gln)
27 d1agxa_
not modelled
29.8
10
Fold: Glutaminase/AsparaginaseSuperfamily: Glutaminase/AsparaginaseFamily: Glutaminase/Asparaginase28 c3qjaA_
not modelled
28.3
12
PDB header: lyaseChain: A: PDB Molecule: indole-3-glycerol phosphate synthase;PDBTitle: crystal structure of the mycobacterium tuberculosis indole-3-glycerol2 phosphate synthase (trpc) in apo form
29 c1wnfA_
not modelled
26.8
13
PDB header: hydrolaseChain: A: PDB Molecule: l-asparaginase;PDBTitle: crystal structure of ph0066 from pyrococcus horikoshii
30 c3cwcB_
not modelled
26.6
13
PDB header: transferaseChain: B: PDB Molecule: putative glycerate kinase 2;PDBTitle: crystal structure of putative glycerate kinase 2 from salmonella2 typhimurium lt2
31 d1vc4a_
not modelled
24.3
26
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes32 c3k2qA_
not modelled
22.7
34
PDB header: transferaseChain: A: PDB Molecule: pyrophosphate-dependent phosphofructokinase;PDBTitle: crystal structure of pyrophosphate-dependent2 phosphofructokinase from marinobacter aquaeolei, northeast3 structural genomics consortium target mqr88
33 c3gfoA_
not modelled
22.2
14
PDB header: atp binding proteinChain: A: PDB Molecule: cobalt import atp-binding protein cbio 1;PDBTitle: structure of cbio1 from clostridium perfringens: part of2 the abc transporter complex cbionq.
34 c2nydB_
not modelled
22.0
7
PDB header: unknown functionChain: B: PDB Molecule: upf0135 protein sa1388;PDBTitle: crystal structure of staphylococcus aureus hypothetical protein sa1388
35 d1wsaa_
not modelled
21.0
15
Fold: Glutaminase/AsparaginaseSuperfamily: Glutaminase/AsparaginaseFamily: Glutaminase/Asparaginase36 c2higA_
not modelled
20.4
18
PDB header: transferaseChain: A: PDB Molecule: 6-phospho-1-fructokinase;PDBTitle: crystal structure of phosphofructokinase apoenzyme from trypanosoma2 brucei.
37 d2axto1
not modelled
19.9
14
Fold: Transmembrane beta-barrelsSuperfamily: OMPA-likeFamily: PsbO-like38 c2x24B_
not modelled
19.4
29
PDB header: ligaseChain: B: PDB Molecule: acetyl-coa carboxylase;PDBTitle: bovine acc2 ct domain in complex with inhibitor
39 c3o3cD_
not modelled
19.4
12
PDB header: transferaseChain: D: PDB Molecule: glycogen [starch] synthase isoform 2;PDBTitle: glycogen synthase basal state udp complex
40 d1g2912
not modelled
18.3
10
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like41 d1js3a_
not modelled
18.2
9
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Pyridoxal-dependent decarboxylase42 c1pixB_
not modelled
16.6
21
PDB header: lyaseChain: B: PDB Molecule: glutaconyl-coa decarboxylase a subunit;PDBTitle: crystal structure of the carboxyltransferase subunit of the2 bacterial ion pump glutaconyl-coenzyme a decarboxylase
43 d1jj7a_
not modelled
16.6
12
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like44 d3d31a2
not modelled
15.4
10
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like45 c3n6rF_
not modelled
15.2
20
PDB header: ligaseChain: F: PDB Molecule: propionyl-coa carboxylase, beta subunit;PDBTitle: crystal structure of the holoenzyme of propionyl-coa carboxylase (pcc)
46 c2yvaB_
not modelled
14.9
19
PDB header: dna binding proteinChain: B: PDB Molecule: dnaa initiator-associating protein diaa;PDBTitle: crystal structure of escherichia coli diaa
47 d1v43a3
not modelled
14.6
2
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like48 c3o8oC_
not modelled
14.3
10
PDB header: transferaseChain: C: PDB Molecule: 6-phosphofructokinase subunit alpha;PDBTitle: structure of phosphofructokinase from saccharomyces cerevisiae
49 d1oxxk2
not modelled
14.0
15
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like50 c3glmD_
not modelled
14.0
19
PDB header: lyaseChain: D: PDB Molecule: glutaconyl-coa decarboxylase subunit a;PDBTitle: glutaconyl-coa decarboxylase a subunit from clostridium2 symbiosum co-crystallized with crotonyl-coa
51 c1f0xA_
not modelled
13.9
11
PDB header: oxidoreductaseChain: A: PDB Molecule: d-lactate dehydrogenase;PDBTitle: crystal structure of d-lactate dehydrogenase, a peripheral2 membrane respiratory enzyme.
52 d1pfka_
not modelled
13.9
9
Fold: PhosphofructokinaseSuperfamily: PhosphofructokinaseFamily: Phosphofructokinase53 d1uyra1
not modelled
13.8
21
Fold: ClpP/crotonaseSuperfamily: ClpP/crotonaseFamily: Biotin dependent carboxylase carboxyltransferase domain54 c1vrgE_
not modelled
13.4
22
PDB header: ligaseChain: E: PDB Molecule: propionyl-coa carboxylase, beta subunit;PDBTitle: crystal structure of propionyl-coa carboxylase, beta subunit (tm0716)2 from thermotoga maritima at 2.30 a resolution
55 d2zcta1
not modelled
13.1
14
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like56 d1i4na_
not modelled
13.0
18
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes57 c2f9yB_
not modelled
12.8
27
PDB header: ligaseChain: B: PDB Molecule: acetyl-coenzyme a carboxylase carboxyl transferase subunitPDBTitle: the crystal structure of the carboxyltransferase subunit of acc from2 escherichia coli
58 d2f9yb1
not modelled
12.8
27
Fold: ClpP/crotonaseSuperfamily: ClpP/crotonaseFamily: Biotin dependent carboxylase carboxyltransferase domain59 d1sffa_
not modelled
12.5
12
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: GABA-aminotransferase-like60 c1x0uB_
not modelled
12.2
22
PDB header: lyaseChain: B: PDB Molecule: hypothetical methylmalonyl-coa decarboxylase alpha subunit;PDBTitle: crystal structure of the carboxyl transferase subunit of putative pcc2 of sulfolobus tokodaii
61 c3eurA_
not modelled
12.0
0
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of the c-terminal domain of uncharacterized protein2 from bacteroides fragilis nctc 9343
62 d2acfa1
not modelled
11.8
10
Fold: Macro domain-likeSuperfamily: Macro domain-likeFamily: Macro domain63 c1drwA_
not modelled
11.8
12
PDB header: oxidoreductaseChain: A: PDB Molecule: dihydrodipicolinate reductase;PDBTitle: escherichia coli dhpr/nhdh complex
64 c3ogfA_
not modelled
11.6
22
PDB header: de novo proteinChain: A: PDB Molecule: de novo designed dimeric trefoil-fold sub-domain whichPDBTitle: crystal structure of difoil-4p homo-trimer: de novo designed dimeric2 trefoil-fold sub-domain which forms homo-trimer assembly
65 c3u9rB_
not modelled
11.6
26
PDB header: ligaseChain: B: PDB Molecule: methylcrotonyl-coa carboxylase, beta-subunit;PDBTitle: crystal structure of p. aeruginosa 3-methylcrotonyl-coa carboxylase2 (mcc), beta subunit
66 d4pfka_
not modelled
11.5
20
Fold: PhosphofructokinaseSuperfamily: PhosphofructokinaseFamily: Phosphofructokinase67 c1xnwD_
not modelled
11.5
24
PDB header: ligaseChain: D: PDB Molecule: propionyl-coa carboxylase complex b subunit;PDBTitle: acyl-coa carboxylase beta subunit from s. coelicolor (pccb),2 apo form #2, mutant d422i
68 c3trjC_
not modelled
11.4
20
PDB header: isomeraseChain: C: PDB Molecule: phosphoheptose isomerase;PDBTitle: structure of a phosphoheptose isomerase from francisella tularensis
69 d1l2ta_
not modelled
11.3
10
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like70 d1vrga1
not modelled
11.1
22
Fold: ClpP/crotonaseSuperfamily: ClpP/crotonaseFamily: Biotin dependent carboxylase carboxyltransferase domain71 c2a7sD_
not modelled
11.1
27
PDB header: ligaseChain: D: PDB Molecule: probable propionyl-coa carboxylase beta chain 5;PDBTitle: crystal structure of the acyl-coa carboxylase, accd5, from2 mycobacterium tuberculosis
72 c2yyzA_
not modelled
10.9
12
PDB header: transport proteinChain: A: PDB Molecule: sugar abc transporter, atp-binding protein;PDBTitle: crystal structure of sugar abc transporter, atp-binding protein
73 c1od4C_
not modelled
10.7
21
PDB header: ligaseChain: C: PDB Molecule: acetyl-coenzyme a carboxylase;PDBTitle: acetyl-coa carboxylase carboxyltransferase domain
74 c3fvqB_
not modelled
10.4
18
PDB header: hydrolaseChain: B: PDB Molecule: fe(3+) ions import atp-binding protein fbpc;PDBTitle: crystal structure of the nucleotide binding domain fbpc2 complexed with atp
75 d1xnya1
not modelled
10.3
24
Fold: ClpP/crotonaseSuperfamily: ClpP/crotonaseFamily: Biotin dependent carboxylase carboxyltransferase domain76 c3ewlA_
not modelled
10.3
0
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized conserved protein bf1870;PDBTitle: crystal structure of conserved protein bf1870 of unknown function from2 bacteroides fragilis
77 c2nq2C_
not modelled
10.2
16
PDB header: metal transportChain: C: PDB Molecule: hypothetical abc transporter atp-binding proteinPDBTitle: an inward-facing conformation of a putative metal-chelate2 type abc transporter.
78 c1z47B_
not modelled
10.2
7
PDB header: ligand binding proteinChain: B: PDB Molecule: putative abc-transporter atp-binding protein;PDBTitle: structure of the atpase subunit cysa of the putative2 sulfate atp-binding cassette (abc) transporter from3 alicyclobacillus acidocaldarius
79 d1nmpa_
not modelled
10.0
12
Fold: NIF3 (NGG1p interacting factor 3)-likeSuperfamily: NIF3 (NGG1p interacting factor 3)-likeFamily: NIF3 (NGG1p interacting factor 3)-like80 d1on3a1
not modelled
10.0
29
Fold: ClpP/crotonaseSuperfamily: ClpP/crotonaseFamily: Biotin dependent carboxylase carboxyltransferase domain81 c2d62A_
not modelled
9.9
9
PDB header: sugar binding proteinChain: A: PDB Molecule: multiple sugar-binding transport atp-bindingPDBTitle: crystal structure of multiple sugar binding transport atp-2 binding protein
82 c3d34A_
not modelled
9.8
44
PDB header: immune systemChain: A: PDB Molecule: spondin-2;PDBTitle: structure of the f-spondin domain of mindin
83 c3opyH_
not modelled
9.2
20
PDB header: transferaseChain: H: PDB Molecule: 6-phosphofructo-1-kinase beta-subunit;PDBTitle: crystal structure of pichia pastoris phosphofructokinase in the t-2 state
84 c3opyB_
not modelled
9.2
20
PDB header: transferaseChain: B: PDB Molecule: 6-phosphofructo-1-kinase beta-subunit;PDBTitle: crystal structure of pichia pastoris phosphofructokinase in the t-2 state
85 d1sgwa_
not modelled
9.0
14
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like86 d2b7ka1
not modelled
9.0
9
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like87 c1m56G_
not modelled
8.8
7
PDB header: oxidoreductaseChain: G: PDB Molecule: cytochrome c oxidase;PDBTitle: structure of cytochrome c oxidase from rhodobactor2 sphaeroides (wild type)
88 c3dhwC_
not modelled
8.8
14
PDB header: membrane protein/hydrolaseChain: C: PDB Molecule: methionine import atp-binding protein metn;PDBTitle: crystal structure of methionine importer metni
89 c1uytC_
not modelled
8.7
21
PDB header: transferaseChain: C: PDB Molecule: acetyl-coa carboxylase;PDBTitle: acetyl-coa carboxylase carboxyltransferase domain
90 c1piiA_
not modelled
8.5
10
PDB header: bifunctional(isomerase and synthase)Chain: A: PDB Molecule: n-(5'phosphoribosyl)anthranilate isomerase;PDBTitle: three-dimensional structure of the bifunctional enzyme2 phosphoribosylanthranilate isomerase:3 indoleglycerolphosphate synthase from escherichia coli4 refined at 2.0 angstroms resolution
91 c3h0jA_
not modelled
8.5
21
PDB header: transferaseChain: A: PDB Molecule: acetyl-coa carboxylase;PDBTitle: crystal structure of the carboxyltransferase domain of2 acetyl-coenzyme a carboxylase in complex with compound 2
92 c3opyG_
not modelled
8.3
10
PDB header: transferaseChain: G: PDB Molecule: 6-phosphofructo-1-kinase alpha-subunit;PDBTitle: crystal structure of pichia pastoris phosphofructokinase in the t-2 state
93 d1ohwa_
not modelled
8.3
14
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: GABA-aminotransferase-like94 c3ff6D_
not modelled
8.3
26
PDB header: ligaseChain: D: PDB Molecule: acetyl-coa carboxylase 2;PDBTitle: human acc2 ct domain with cp-640186
95 d1xrsa_
not modelled
8.1
20
Fold: TIM beta/alpha-barrelSuperfamily: Cobalamin (vitamin B12)-dependent enzymesFamily: D-lysine 5,6-aminomutase alpha subunit, KamD96 d2awna2
not modelled
8.0
10
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like97 c2p31B_
not modelled
8.0
11
PDB header: oxidoreductaseChain: B: PDB Molecule: glutathione peroxidase 7;PDBTitle: crystal structure of human glutathione peroxidase 7
98 c2it1B_
not modelled
8.0
10
PDB header: transport proteinChain: B: PDB Molecule: 362aa long hypothetical maltose/maltodextrinPDBTitle: structure of ph0203 protein from pyrococcus horikoshii
99 d1prxa_
not modelled
7.8
23
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like