| View Alignment |
Fold Library |
Template Length |
Model
| PSSM E_value |
SAWTED E-value |
Biotext |
Class |
Fold |
SuperFamily |
Family |
Protein |
Species |
ID |
Region |
CS |
PSSM Type
|
 |
d1tfr_1 19%i.d. |
123 |
|
0.111 |
1 | n/a |
All alpha proteins |
SAM domain-like |
5' to 3' exonuclease, C-terminal subdomain |
5' to 3' exonuclease, C-terminal subdomain |
T4 RNase H |
Bacteriophage T4 |
1tfr |
183-305
|
0.802 |
1D |
 |
d1higa_ 11%i.d. |
123 |
|
2.49 |
0.885

| 1.00
|
All alpha proteins |
4-helical cytokines |
4-helical cytokines |
Interferons/interleukin-10 (IL-10) |
Interferon-gamma |
Human (Homo sapiens) |
1hig |
a:
|
0.500 |
3D |
 |
d1iara_ 12%i.d. |
129 |

|
3.16 |
0.751

| 1.00
|
All alpha proteins |
4-helical cytokines |
4-helical cytokines |
Short-chain cytokines |
Interleukin-4 (IL-4) |
Human (Homo sapiens) |
1iar |
a:
|
0.500 |
R1D |
 |
d1vola1 17%i.d. |
95 |
|
4.67 |
0.792

| 1.00
|
All alpha proteins |
Cyclin-like |
Cyclin-like |
Transcription factor IIB (TFIIB), core domain |
Transcription factor IIB (TFIIB), core domain |
Human (Homo sapiens) |
1vol |
a:113-207
|
0.943 |
1D |
 |
c1g4yb_ 16%i.d. |
81 |
|
5.09 |
1 | 1.00
|
not in SCOP 1.53 |
PDB header: signaling protein. |
Chain: B: PDB Molecule:calcium-activated potassium channel rsk2;. |
. |
PDBTitle: 1.60 a crystal structure of the gating domain from small2 conductance potassium channel complexed with calcium-3 calmodulin
.
| - |
1g4y |
b
|
0.000 |
1D |
 |
c1ytfd_ 17%i.d. |
100 |
|
5.4 |
0.82

| 1.00
|
not in SCOP 1.53 |
PDB header: complex (transcription regulation/dna). |
Chain: D: PDB Molecule:yeast transcription factor iia;. |
. |
PDBTitle: yeast tfiia/tbp/dna complex
.
| - |
1ytf |
d
|
0.000 |
1D |
 |
d1fipa_ 22%i.d. |
73 |

|
7.01 |
1 | 1.00
|
All alpha proteins |
FIS-like |
FIS-like |
FIS-like |
FIS protein |
Escherichia coli |
1fip |
a:
|
0.000 |
1D |
 |
d1d9ca_ 11%i.d. |
121 |
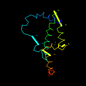
|
7.74 |
0.87

| 1.00
|
All alpha proteins |
4-helical cytokines |
4-helical cytokines |
Interferons/interleukin-10 (IL-10) |
Interferon-gamma |
Bovine (Bos taurus) |
1d9c |
a:
|
0.500 |
1D |
 |
c1jm7b_ 13%i.d. |
97 |
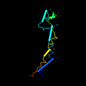
|
8.5 |
1 | 1.00
|
not in SCOP 1.53 |
PDB header: antitumor. |
Chain: B: PDB Molecule:brca1-associated ring domain protein 1;. |
. |
PDBTitle: solution structure of the brca1/bard1 ring-domain2 heterodimer
.
| - |
1jm7 |
b
|
0.000 |
R1D |
 |
c1gjha_ 18%i.d. |
164 |

|
8.51 |
0.631

| 1.00
|
Membrane and cell surface proteins and peptides |
Toxins' membrane translocation domains |
Bcl-2 inhibitors of programmed cell death |
homol. to d1maz__ |
- |
- |
1gjh |
a
|
0.000 |
1D |
 |
d2rig__ 14%i.d. |
119 |
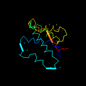
|
8.62 |
0.869

| 1.00
|
All alpha proteins |
4-helical cytokines |
4-helical cytokines |
Interferons/interleukin-10 (IL-10) |
Interferon-gamma |
Rabbit (Oryctolagus cuniculus) |
2rig |
-
|
0.500 |
1D |
 |
d1brwa1 10%i.d. |
70 |

|
8.77 |
1 | 1.00
|
All alpha proteins |
Methionine synthase domain-like |
Pyrimidine nucleoside phosphorylase N-terminal domain |
Pyrimidine nucleoside phosphorylase N-terminal domain |
Pyrimidine nucleoside phosphorylase |
Bacillus stearothermophilus |
1brw |
a:1-70
|
0.000 |
3D |
 |
d1af7_1 12%i.d. |
81 |

|
9.05 |
1 | 1.00
|
All alpha proteins |
Chemotaxis receptor methyltransferase CheR, N-terminal domain |
Chemotaxis receptor methyltransferase CheR, N-terminal domain |
Chemotaxis receptor methyltransferase CheR, N-terminal domain |
Chemotaxis receptor methyltransferase CheR, N-terminal domain |
Salmonella typhimurium |
1af7 |
11-91
|
0.500 |
R1D |
 |
d1a77_1 12%i.d. |
108 |
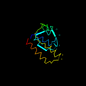
|
10 |
1 | 1.00
|
All alpha proteins |
SAM domain-like |
5' to 3' exonuclease, C-terminal subdomain |
5' to 3' exonuclease, C-terminal subdomain |
Flap endonuclease-1 |
Methanococcus jannaschii |
1a77 |
209-316
|
0.912 |
1D |
 |
d1aisb1 7%i.d. |
98 |
|
10.3 |
0.829

| 1.00
|
All alpha proteins |
Cyclin-like |
Cyclin-like |
Transcription factor IIB (TFIIB), core domain |
Transcription factor IIB (TFIIB), core domain |
Pyrococcus woesei |
1ais |
b:1108-1205
|
0.968 |
3D |
 |
c1e6ia_ 19%i.d. |
110 |

|
10.4 |
1 | 1.00
|
All alpha proteins |
Bromodomain-like |
Bromodomain |
homol. to d1b91a_ |
- |
- |
1e6i |
a
|
0.000 |
1D |
 |
d1a0p_1 12%i.d. |
91 |

|
10.6 |
1 | 1.00
|
All alpha proteins |
SAM domain-like |
lambda integrase-like, N-terminal domain |
lambda integrase-like, N-terminal domain |
Recombinase XerD |
Escherichia coli |
1a0p |
3-100
|
0.000 |
R1D |
 |
d1vin_1 20%i.d. |
128 |
|
10.6 |
1 | 1.00
|
All alpha proteins |
Cyclin-like |
Cyclin-like |
Cyclin |
Cyclin A |
Bovine (Bos taurus) |
1vin |
181-308
|
1.000 |
1D |
 |
c1gd8a_ 12%i.d. |
105 |

|
11 |
1 | 1.00
|
not in SCOP 1.53 |
PDB header: ribosome. |
Chain: A: PDB Molecule:50s ribosomal protein l17;. |
. |
PDBTitle: the crystal structure of bacteria-specific l17 ribosomal2 protein.
.
| - |
1gd8 |
a
|
0.000 |
R1D |
 |
d1bu2a1 13%i.d. |
127 |

|
11.1 |
1 | 1.00
|
All alpha proteins |
Cyclin-like |
Cyclin-like |
Cyclin |
Viral cyclin |
Herpes virus saimiri |
1bu2 |
a:22-148
|
0.847 |
3D |