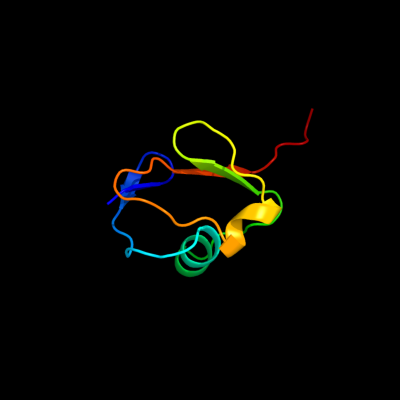
| 1 | c2kl0A_
|
|
 |
99.9 |
29 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative thiamin biosynthesis this;
PDBTitle: solution nmr structure of rhodopseudomonas palustris rpa3574,2 northeast structural genomics consortium (nesg) target rpr325
|
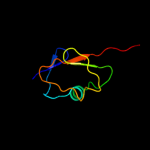
| 2 | d1zud21
|
|
 |
99.9 |
100 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:MoaD/ThiS
Family:ThiS |
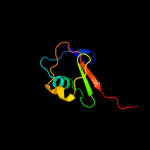
| 3 | c3cwiA_
|
|
 |
99.8 |
24 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:thiamine-biosynthesis protein this;
PDBTitle: crystal structure of thiamine biosynthesis protein (this)2 from geobacter metallireducens. northeast structural3 genomics consortium target gmr137
|
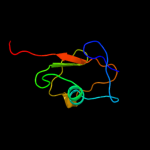
| 4 | d2cu3a1
|
|
 |
99.8 |
21 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:MoaD/ThiS
Family:ThiS |
| 5 | d1tygb_
|
|
 |
99.8 |
17 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:MoaD/ThiS
Family:ThiS |
| 6 | c1tygG_
|
|
 |
99.8 |
16 |
PDB header:biosynthetic protein
Chain: G: PDB Molecule:yjbs;
PDBTitle: structure of the thiazole synthase/this complex
|
| 7 | c2g1eA_
|
|
 |
98.2 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:hypothetical protein ta0895;
PDBTitle: solution structure of ta0895
|
| 8 | c2qieB_
|
|
 |
98.2 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:molybdopterin synthase small subunit;
PDBTitle: staphylococcus aureus molybdopterin synthase in complex2 with precursor z
|
| 9 | c3po0A_
|
|
 |
97.9 |
22 |
PDB header:protein binding
Chain: A: PDB Molecule:small archaeal modifier protein 1;
PDBTitle: crystal structure of samp1 from haloferax volcanii
|
| 10 | d1v8ca1
|
|
 |
97.8 |
25 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:MoaD/ThiS
Family:MoaD |
| 11 | c3rpfC_
|
|
 |
97.7 |
26 |
PDB header:transferase
Chain: C: PDB Molecule:molybdopterin converting factor, subunit 1 (moad);
PDBTitle: protein-protein complex of subunit 1 and 2 of molybdopterin-converting2 factor from helicobacter pylori 26695
|
| 12 | d1fm0d_
|
|
 |
97.6 |
28 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:MoaD/ThiS
Family:MoaD |
| 13 | c2qjlA_
|
|
 |
97.6 |
26 |
PDB header:signaling protein
Chain: A: PDB Molecule:ubiquitin-related modifier 1;
PDBTitle: crystal structure of urm1
|
| 14 | c2l52A_
|
|
 |
97.5 |
20 |
PDB header:protein binding
Chain: A: PDB Molecule:methanosarcina acetivorans samp1 homolog;
PDBTitle: solution structure of the small archaeal modifier protein 1 (samp1)2 from methanosarcina acetivorans
|
| 15 | d1xo3a_
|
|
 |
97.4 |
30 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:MoaD/ThiS
Family:C9orf74 homolog |
| 16 | d1vjka_
|
|
 |
97.4 |
23 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:MoaD/ThiS
Family:MoaD |
| 17 | c2k9xA_
|
|
 |
97.4 |
21 |
PDB header:unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution structure of urm1 from trypanosoma brucei
|
| 18 | d1wgka_
|
|
 |
97.3 |
24 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:MoaD/ThiS
Family:C9orf74 homolog |
| 19 | c1v8cA_
|
|
 |
97.3 |
28 |
PDB header:protein binding
Chain: A: PDB Molecule:moad related protein;
PDBTitle: crystal structure of moad related protein from thermus2 thermophilus hb8
|
| 20 | c3dwmA_
|
|
 |
97.2 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:9.5 kda culture filtrate antigen cfp10a;
PDBTitle: crystal structure of mycobacterium tuberculosis cyso, an antigen
|
| 21 | d1ryja_ |
|
not modelled |
97.2 |
29 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:MoaD/ThiS
Family:ThiS |
| 22 | c2kmmA_ |
|
not modelled |
97.0 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:guanosine-3',5'-bis(diphosphate) 3'-
PDBTitle: solution nmr structure of the tgs domain of pg1808 from2 porphyromonas gingivalis. northeast structural genomics3 consortium target pgr122a (418-481)
|
| 23 | d1rwsa_ |
|
not modelled |
96.9 |
24 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:MoaD/ThiS
Family:ThiS |
| 24 | c3hvzB_ |
|
not modelled |
96.1 |
17 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of the tgs domain of the clolep_03100 protein from2 clostridium leptum, northeast structural genomics consortium target3 qlr13a
|
| 25 | d1tkea1 |
|
not modelled |
95.3 |
11 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:TGS-like
Family:TGS domain |
| 26 | d3c8ya2 |
|
not modelled |
94.9 |
13 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:2Fe-2S ferredoxin-like
Family:2Fe-2S ferredoxin domains from multidomain proteins |
| 27 | d1vlba2 |
|
not modelled |
94.8 |
17 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:2Fe-2S ferredoxin-like
Family:2Fe-2S ferredoxin domains from multidomain proteins |
| 28 | c1wwtA_ |
|
not modelled |
94.4 |
5 |
PDB header:ligase
Chain: A: PDB Molecule:threonyl-trna synthetase, cytoplasmic;
PDBTitle: solution structure of the tgs domain from human threonyl-2 trna synthetase
|
| 29 | c1c4cA_ |
|
not modelled |
93.6 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein (fe-only hydrogenase);
PDBTitle: binding of exogenously added carbon monoxide at the active2 site of the fe-only hydrogenase (cpi) from clostridium3 pasteurianum
|
| 30 | c2hj1A_ |
|
not modelled |
93.5 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of a 3d domain-swapped dimer of protein hi0395 from2 haemophilus influenzae
|
| 31 | d2hj1a1 |
|
not modelled |
93.5 |
18 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:MoaD/ThiS
Family:HI0395-like |
| 32 | c1nyqA_ |
|
not modelled |
92.6 |
17 |
PDB header:ligase
Chain: A: PDB Molecule:threonyl-trna synthetase 1;
PDBTitle: structure of staphylococcus aureus threonyl-trna synthetase2 complexed with an analogue of threonyl adenylate
|
| 33 | d1dgja2 |
|
not modelled |
91.9 |
9 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:2Fe-2S ferredoxin-like
Family:2Fe-2S ferredoxin domains from multidomain proteins |
| 34 | d1t3qa2 |
|
not modelled |
91.4 |
10 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:2Fe-2S ferredoxin-like
Family:2Fe-2S ferredoxin domains from multidomain proteins |
| 35 | d1n62a2 |
|
not modelled |
91.2 |
11 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:2Fe-2S ferredoxin-like
Family:2Fe-2S ferredoxin domains from multidomain proteins |
| 36 | d1ffva2 |
|
not modelled |
89.9 |
17 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:2Fe-2S ferredoxin-like
Family:2Fe-2S ferredoxin domains from multidomain proteins |
| 37 | d2fug33 |
|
not modelled |
89.7 |
13 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:2Fe-2S ferredoxin-like
Family:2Fe-2S ferredoxin domains from multidomain proteins |
| 38 | d1rm6c2 |
|
not modelled |
89.0 |
7 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:2Fe-2S ferredoxin-like
Family:2Fe-2S ferredoxin domains from multidomain proteins |
| 39 | d1ud7a_ |
|
not modelled |
88.0 |
15 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:Ubiquitin-related |
| 40 | d1wxqa2 |
|
not modelled |
88.0 |
19 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:TGS-like
Family:G domain-linked domain |
| 41 | c1tkeA_ |
|
not modelled |
87.8 |
11 |
PDB header:ligase
Chain: A: PDB Molecule:threonyl-trna synthetase;
PDBTitle: crystal structure of the editing domain of threonyl-trna2 synthetase complexed with serine
|
| 42 | c1yx5B_ |
|
not modelled |
86.1 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:ubiquitin;
PDBTitle: solution structure of s5a uim-1/ubiquitin complex
|
| 43 | d1c3ta_ |
|
not modelled |
85.7 |
17 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:Ubiquitin-related |
| 44 | c2ekiA_ |
|
not modelled |
84.2 |
15 |
PDB header:signaling protein
Chain: A: PDB Molecule:developmentally-regulated gtp-binding protein 1;
PDBTitle: solution structures of the tgs domain of human2 developmentally-regulated gtp-binding protein 1
|
| 45 | c1n60D_ |
|
not modelled |
83.8 |
12 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:carbon monoxide dehydrogenase small chain;
PDBTitle: crystal structure of the cu,mo-co dehydrogenase (codh); cyanide-2 inactivated form
|
| 46 | c1t3qD_ |
|
not modelled |
82.9 |
7 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:quinoline 2-oxidoreductase small subunit;
PDBTitle: crystal structure of quinoline 2-oxidoreductase from pseudomonas2 putida 86
|
| 47 | d1nyra2 |
|
not modelled |
80.7 |
16 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:TGS-like
Family:TGS domain |
| 48 | c2fugC_ |
|
not modelled |
79.1 |
13 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:nadh-quinone oxidoreductase chain 3;
PDBTitle: crystal structure of the hydrophilic domain of respiratory complex i2 from thermus thermophilus
|
| 49 | c1ffuA_ |
|
not modelled |
78.7 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:cuts, iron-sulfur protein of carbon monoxide
PDBTitle: carbon monoxide dehydrogenase from hydrogenophaga2 pseudoflava which lacks the mo-pyranopterin moiety of the3 molybdenum cofactor
|
| 50 | c3hrdH_ |
|
not modelled |
78.5 |
15 |
PDB header:oxidoreductase
Chain: H: PDB Molecule:nicotinate dehydrogenase small fes subunit;
PDBTitle: crystal structure of nicotinate dehydrogenase
|
| 51 | d1euvb_ |
|
not modelled |
76.3 |
12 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:Ubiquitin-related |
| 52 | c1rm6F_ |
|
not modelled |
75.5 |
7 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:4-hydroxybenzoyl-coa reductase gamma subunit;
PDBTitle: structure of 4-hydroxybenzoyl-coa reductase from thauera2 aromatica
|
| 53 | c1qf6A_ |
|
not modelled |
73.4 |
13 |
PDB header:ligase/rna
Chain: A: PDB Molecule:threonyl-trna synthetase;
PDBTitle: structure of e. coli threonyl-trna synthetase complexed with its2 cognate trna
|
| 54 | d1uela_ |
|
not modelled |
72.8 |
18 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:Ubiquitin-related |
| 55 | c1vlbA_ |
|
not modelled |
72.8 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aldehyde oxidoreductase;
PDBTitle: structure refinement of the aldehyde oxidoreductase from2 desulfovibrio gigas at 1.28 a
|
| 56 | d1ogwa_ |
|
not modelled |
72.1 |
12 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:Ubiquitin-related |
| 57 | d1wm3a_ |
|
not modelled |
71.5 |
17 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:Ubiquitin-related |
| 58 | c2kd0A_ |
|
not modelled |
70.9 |
20 |
PDB header:signaling protein
Chain: A: PDB Molecule:lrr repeats and ubiquitin-like domain-containing
PDBTitle: nmr solution structure of o64736 protein from arabidopsis2 thaliana. northeast structural genomics consortium mega3 target ar3445a
|
| 59 | d1wgga_ |
|
not modelled |
69.7 |
12 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:Ubiquitin-related |
| 60 | d1jroa2 |
|
not modelled |
69.4 |
21 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:2Fe-2S ferredoxin-like
Family:2Fe-2S ferredoxin domains from multidomain proteins |
| 61 | d1v97a2 |
|
not modelled |
69.4 |
17 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:2Fe-2S ferredoxin-like
Family:2Fe-2S ferredoxin domains from multidomain proteins |
| 62 | d1v86a_ |
|
not modelled |
66.5 |
15 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:Ubiquitin-related |
| 63 | c3tixA_ |
|
not modelled |
64.4 |
12 |
PDB header:gene regulation/protein binding
Chain: A: PDB Molecule:ubiquitin-like protein smt3, rna-induced transcriptional
PDBTitle: crystal structure of the chp1-tas3 complex core
|
| 64 | d1bt0a_ |
|
not modelled |
63.2 |
12 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:Ubiquitin-related |
| 65 | c3a4rB_ |
|
not modelled |
63.0 |
15 |
PDB header:transcription
Chain: B: PDB Molecule:nfatc2-interacting protein;
PDBTitle: the crystal structure of sumo-like domain 2 in nip45
|
| 66 | d2io3b1 |
|
not modelled |
60.7 |
21 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:Ubiquitin-related |
| 67 | d1we7a_ |
|
not modelled |
59.8 |
14 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:Ubiquitin-related |
| 68 | d1ndda_ |
|
not modelled |
59.3 |
13 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:Ubiquitin-related |
| 69 | c1dgjA_ |
|
not modelled |
59.3 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aldehyde oxidoreductase;
PDBTitle: crystal structure of the aldehyde oxidoreductase from2 desulfovibrio desulfuricans atcc 27774
|
| 70 | c2jxxA_ |
|
not modelled |
58.1 |
13 |
PDB header:protein binding
Chain: A: PDB Molecule:nfatc2-interacting protein;
PDBTitle: nmr solution structure of ubiquitin-like domain of2 nfatc2ip. northeast structural genomics consortium target3 hr5627
|
| 71 | d1wh3a_ |
|
not modelled |
57.8 |
17 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:Ubiquitin-related |
| 72 | c3pgeA_ |
|
not modelled |
57.1 |
12 |
PDB header:dna binding protein
Chain: A: PDB Molecule:sumo-modified proliferating cell nuclear antigen;
PDBTitle: structure of sumoylated pcna
|
| 73 | c3goeA_ |
|
not modelled |
54.6 |
17 |
PDB header:recombination, replication
Chain: A: PDB Molecule:dna repair protein rad60;
PDBTitle: molecular mimicry of sumo promotes dna repair
|
| 74 | c3eubJ_ |
|
not modelled |
53.7 |
18 |
PDB header:oxidoreductase
Chain: J: PDB Molecule:xanthine dehydrogenase/oxidase;
PDBTitle: crystal structure of desulfo-xanthine oxidase with xanthine
|
| 75 | c3b9jI_ |
|
not modelled |
52.8 |
18 |
PDB header:oxidoreductase
Chain: I: PDB Molecule:xanthine oxidase;
PDBTitle: structure of xanthine oxidase with 2-hydroxy-6-methylpurine
|
| 76 | d2faza1 |
|
not modelled |
52.6 |
18 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:Ubiquitin-related |
| 77 | d1sifa_ |
|
not modelled |
52.5 |
10 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:Ubiquitin-related |
| 78 | c2kanA_ |
|
not modelled |
51.6 |
12 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein ar3433a;
PDBTitle: solution nmr structure of ubiquitin-like domain of2 arabidopsis thaliana protein at2g32350. northeast3 structural genomics consortium target ar3433a
|
| 79 | d2uyzb1 |
|
not modelled |
51.6 |
12 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:Ubiquitin-related |
| 80 | c2k8hA_ |
|
not modelled |
51.2 |
10 |
PDB header:signaling protein
Chain: A: PDB Molecule:small ubiquitin protein;
PDBTitle: solution structure of sumo from trypanosoma brucei
|
| 81 | c2ekeC_ |
|
not modelled |
51.1 |
10 |
PDB header:ligase/protein binding
Chain: C: PDB Molecule:ubiquitin-like protein smt3;
PDBTitle: structure of a sumo-binding-motif mimic bound to smt3p-2 ubc9p: conservation of a noncovalent ubiquitin-like3 protein-e2 complex as a platform for selective4 interactions within a sumo pathway
|
| 82 | d2piea1 |
|
not modelled |
47.7 |
21 |
Fold:SMAD/FHA domain
Superfamily:SMAD/FHA domain
Family:FHA domain |
| 83 | d2g1la1 |
|
not modelled |
47.3 |
23 |
Fold:SMAD/FHA domain
Superfamily:SMAD/FHA domain
Family:FHA domain |
| 84 | d1a5ra_ |
|
not modelled |
46.6 |
14 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:Ubiquitin-related |
| 85 | d1gxca_ |
|
not modelled |
46.6 |
17 |
Fold:SMAD/FHA domain
Superfamily:SMAD/FHA domain
Family:FHA domain |
| 86 | c1gxcA_ |
|
not modelled |
46.6 |
17 |
PDB header:phosphoprotein-binding domain
Chain: A: PDB Molecule:serine/threonine-protein kinase chk2;
PDBTitle: fha domain from human chk2 kinase in complex with a2 synthetic phosphopeptide
|
| 87 | c2l76A_ |
|
not modelled |
46.4 |
9 |
PDB header:transcription
Chain: A: PDB Molecule:nfatc2-interacting protein;
PDBTitle: solution nmr structure of human nfatc2ip ubiquitin-like domain,2 nfatc2ip_244_338, nesg target ht65a/ocsp target hs00387_244_338/sgc-3 toronto
|
| 88 | d1wz0a1 |
|
not modelled |
45.4 |
17 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:Ubiquitin-related |
| 89 | d1v5oa_ |
|
not modelled |
44.1 |
15 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:Ubiquitin-related |
| 90 | d1yfba1 |
|
not modelled |
44.0 |
14 |
Fold:Double-split beta-barrel
Superfamily:AbrB/MazE/MraZ-like
Family:AbrB N-terminal domain-like |
| 91 | c2l7rA_ |
|
not modelled |
43.3 |
15 |
PDB header:protein binding
Chain: A: PDB Molecule:ubiquitin-like protein fubi;
PDBTitle: solution nmr structure of n-terminal ubiquitin-like domain of fubi, a2 ribosomal protein s30 precursor from homo sapiens. northeast3 structural genomics consortium (nesg) target hr6166
|
| 92 | d1j8ca_ |
|
not modelled |
42.8 |
15 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:Ubiquitin-related |
| 93 | c3dh3C_ |
|
not modelled |
42.7 |
12 |
PDB header:isomerase/rna
Chain: C: PDB Molecule:ribosomal large subunit pseudouridine synthase f;
PDBTitle: crystal structure of rluf in complex with a 22 nucleotide2 rna substrate
|
| 94 | d1g3ga_ |
|
not modelled |
42.4 |
25 |
Fold:SMAD/FHA domain
Superfamily:SMAD/FHA domain
Family:FHA domain |
| 95 | c2eh0A_ |
|
not modelled |
41.0 |
18 |
PDB header:transport protein
Chain: A: PDB Molecule:kinesin-like protein kif1b;
PDBTitle: solution structure of the fha domain from human kinesin-2 like protein kif1b
|
| 96 | c3lzkC_ |
|
not modelled |
40.8 |
10 |
PDB header:hydrolase
Chain: C: PDB Molecule:fumarylacetoacetate hydrolase family protein;
PDBTitle: the crystal structure of a probably aromatic amino acid2 degradation protein from sinorhizobium meliloti 1021
|
| 97 | c3kt9A_ |
|
not modelled |
40.3 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:aprataxin;
PDBTitle: aprataxin fha domain
|
| 98 | d2fy9a1 |
|
not modelled |
40.2 |
23 |
Fold:Double-split beta-barrel
Superfamily:AbrB/MazE/MraZ-like
Family:AbrB N-terminal domain-like |
| 99 | d1iyfa_ |
|
not modelled |
40.0 |
7 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:Ubiquitin-related |
| 100 | c3hx1B_ |
|
not modelled |
39.4 |
30 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:slr1951 protein;
PDBTitle: crystal structure of the slr1951 protein from synechocystis sp.2 northeast structural genomics consortium target sgr167a
|
| 101 | c2w3rG_ |
|
not modelled |
38.5 |
18 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:xanthine dehydrogenase;
PDBTitle: crystal structure of xanthine dehydrogenase (desulfo form)2 from rhodobacter capsulatus in complex with hypoxanthine
|
| 102 | d1yjma1 |
|
not modelled |
38.4 |
13 |
Fold:SMAD/FHA domain
Superfamily:SMAD/FHA domain
Family:FHA domain |
| 103 | c3jxoB_ |
|
not modelled |
37.0 |
20 |
PDB header:transport protein
Chain: B: PDB Molecule:trka-n domain protein;
PDBTitle: crystal structure of an octomeric two-subunit trka k+ channel ring2 gating assembly, tm1088a:tm1088b, from thermotoga maritima
|
| 104 | c2w1tB_ |
|
not modelled |
37.0 |
23 |
PDB header:transcription
Chain: B: PDB Molecule:stage v sporulation protein t;
PDBTitle: crystal structure of b. subtilis spovt
|
| 105 | d1wy8a1 |
|
not modelled |
36.8 |
23 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:Ubiquitin-related |
| 106 | d1p9ka_ |
|
not modelled |
36.7 |
18 |
Fold:Alpha-L RNA-binding motif
Superfamily:Alpha-L RNA-binding motif
Family:YbcJ-like |
| 107 | c3fm8A_ |
|
not modelled |
36.4 |
27 |
PDB header:transport protein/hydrolase activator
Chain: A: PDB Molecule:kinesin-like protein kif13b;
PDBTitle: crystal structure of full length centaurin alpha-1 bound with the fha2 domain of kif13b (capri target)
|
| 108 | c2ro5B_ |
|
not modelled |
36.4 |
23 |
PDB header:transcription
Chain: B: PDB Molecule:stage v sporulation protein t;
PDBTitle: rdc-refined solution structure of the n-terminal dna2 recognition domain of the bacillus subtilis transition-3 state regulator spovt
|
| 109 | c2glwA_ |
|
not modelled |
35.3 |
19 |
PDB header:transcription
Chain: A: PDB Molecule:92aa long hypothetical protein;
PDBTitle: the solution structure of phs018 from pyrococcus horikoshii
|
| 110 | d1dm9a_ |
|
not modelled |
33.5 |
24 |
Fold:Alpha-L RNA-binding motif
Superfamily:Alpha-L RNA-binding motif
Family:Heat shock protein 15 kD |
| 111 | c1dm9A_ |
|
not modelled |
33.5 |
24 |
PDB header:structural genomics
Chain: A: PDB Molecule:hypothetical 15.5 kd protein in mrca-pcka
PDBTitle: heat shock protein 15 kd
|
| 112 | d1zkha1 |
|
not modelled |
33.2 |
16 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:Ubiquitin-related |
| 113 | c2bknA_ |
|
not modelled |
33.0 |
16 |
PDB header:membrane protein
Chain: A: PDB Molecule:hypothetical protein ph0236;
PDBTitle: structure analysis of unknown function protein
|
| 114 | c2jqlA_ |
|
not modelled |
31.2 |
26 |
PDB header:cell cycle
Chain: A: PDB Molecule:dna damage response protein kinase dun1;
PDBTitle: nmr structure of the yeast dun1 fha domain in complex with2 a doubly phosphorylated (pt) peptide derived from rad533 scd1
|
| 115 | c3m62B_ |
|
not modelled |
31.1 |
15 |
PDB header:ligase/protein binding
Chain: B: PDB Molecule:uv excision repair protein rad23;
PDBTitle: crystal structure of ufd2 in complex with the ubiquitin-like (ubl)2 domain of rad23
|
| 116 | c2dziA_ |
|
not modelled |
31.0 |
12 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:ubiquitin-like protein 4a;
PDBTitle: 2dzi/solution structure of the n-terminal ubiquitin-like2 domain in human ubiquitin-like protein 4a (gdx)
|
| 117 | c1yj5C_ |
|
not modelled |
28.5 |
14 |
PDB header:transferase
Chain: C: PDB Molecule:5' polynucleotide kinase-3' phosphatase fha domain;
PDBTitle: molecular architecture of mammalian polynucleotide kinase, a dna2 repair enzyme
|
| 118 | c2kdiA_ |
|
not modelled |
27.8 |
10 |
PDB header:signaling protein
Chain: A: PDB Molecule:ubiquitin, vacuolar protein sorting-associated
PDBTitle: solution structure of a ubiquitin/uim fusion protein
|
| 119 | d1vcta2 |
|
not modelled |
27.4 |
16 |
Fold:TrkA C-terminal domain-like
Superfamily:TrkA C-terminal domain-like
Family:TrkA C-terminal domain-like |
| 120 | d1oqya4 |
|
not modelled |
27.4 |
17 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:Ubiquitin-related |