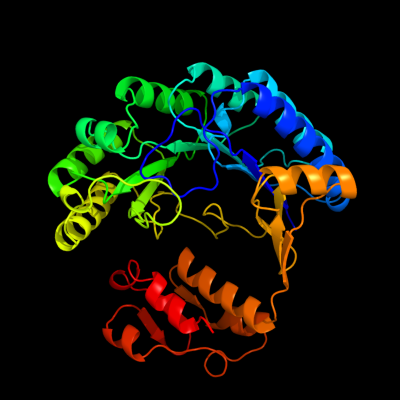
| 1 | d1olta_
|
|
 |
100.0 |
25 |
Fold:TIM beta/alpha-barrel
Superfamily:Radical SAM enzymes
Family:Oxygen-independent coproporphyrinogen III oxidase HemN |
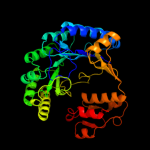
| 2 | c3cixA_
|
|
 |
100.0 |
14 |
PDB header:adomet binding protein
Chain: A: PDB Molecule:fefe-hydrogenase maturase;
PDBTitle: x-ray structure of the [fefe]-hydrogenase maturase hyde from2 thermotoga maritima in complex with thiocyanate
|
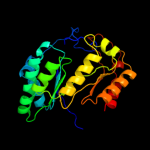
| 3 | c2qgqF_
|
|
 |
99.9 |
14 |
PDB header:structural genomics, unknown function
Chain: F: PDB Molecule:protein tm_1862;
PDBTitle: crystal structure of tm_1862 from thermotoga maritima.2 northeast structural genomics consortium target vr77
|
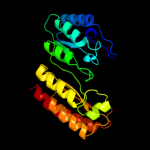
| 4 | c3t7vA_
|
|
 |
99.9 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:methylornithine synthase pylb;
PDBTitle: crystal structure of methylornithine synthase (pylb)
|
| 5 | d1r30a_
|
|
 |
99.9 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Radical SAM enzymes
Family:Biotin synthase |
| 6 | c1r30A_
|
|
 |
99.9 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:biotin synthase;
PDBTitle: the crystal structure of biotin synthase, an s-2 adenosylmethionine-dependent radical enzyme
|
| 7 | c3rfaA_
|
|
 |
99.7 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ribosomal rna large subunit methyltransferase n;
PDBTitle: x-ray structure of rlmn from escherichia coli in complex with s-2 adenosylmethionine
|
| 8 | d1tv8a_
|
|
 |
99.6 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Radical SAM enzymes
Family:MoCo biosynthesis proteins |
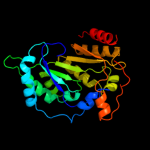
| 9 | c3c8fA_
|
|
 |
99.5 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pyruvate formate-lyase 1-activating enzyme;
PDBTitle: 4fe-4s-pyruvate formate-lyase activating enzyme with2 partially disordered adomet
|
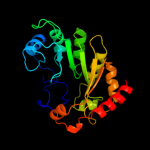
| 10 | c2yx0A_
|
|
 |
99.4 |
13 |
PDB header:metal binding protein
Chain: A: PDB Molecule:radical sam enzyme;
PDBTitle: crystal structure of p. horikoshii tyw1
|
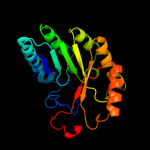
| 11 | c2a5hC_
|
|
 |
99.2 |
14 |
PDB header:isomerase
Chain: C: PDB Molecule:l-lysine 2,3-aminomutase;
PDBTitle: 2.1 angstrom x-ray crystal structure of lysine-2,3-aminomutase from2 clostridium subterminale sb4, with michaelis analog (l-alpha-lysine3 external aldimine form of pyridoxal-5'-phosphate).
|
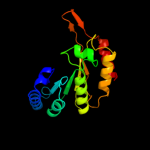
| 12 | c3canA_
|
|
 |
98.3 |
9 |
PDB header:lyase activator
Chain: A: PDB Molecule:pyruvate-formate lyase-activating enzyme;
PDBTitle: crystal structure of a domain of pyruvate-formate lyase-activating2 enzyme from bacteroides vulgatus atcc 8482
|
| 13 | c2z2uA_
|
|
 |
97.8 |
10 |
PDB header:metal binding protein
Chain: A: PDB Molecule:upf0026 protein mj0257;
PDBTitle: crystal structure of archaeal tyw1
|
| 14 | c2x4hA_
|
|
 |
97.0 |
13 |
PDB header:transcription
Chain: A: PDB Molecule:hypothetical protein sso2273;
PDBTitle: crystal structure of the hypothetical protein sso2273 from2 sulfolobus solfataricus
|
| 15 | c2it0A_
|
|
 |
96.7 |
10 |
PDB header:transcription/dna
Chain: A: PDB Molecule:iron-dependent repressor ider;
PDBTitle: crystal structure of a two-domain ider-dna complex crystal2 form ii
|
| 16 | c1f5tA_
|
|
 |
96.6 |
10 |
PDB header:transcription/dna
Chain: A: PDB Molecule:diphtheria toxin repressor;
PDBTitle: diphtheria tox repressor (c102d mutant) complexed with2 nickel and dtxr consensus binding sequence
|
| 17 | c2h09A_
|
|
 |
96.6 |
17 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator mntr;
PDBTitle: crystal structure of diphtheria toxin repressor like protein2 from e. coli
|
| 18 | c1g3wA_
|
|
 |
96.5 |
8 |
PDB header:gene regulation
Chain: A: PDB Molecule:diphtheria toxin repressor;
PDBTitle: cd-cys102ser dtxr
|
| 19 | c1fx7C_
|
|
 |
96.3 |
12 |
PDB header:signaling protein
Chain: C: PDB Molecule:iron-dependent repressor ider;
PDBTitle: crystal structure of the iron-dependent regulator (ider)2 from mycobacterium tuberculosis
|
| 20 | d1r7ja_
|
|
 |
96.2 |
9 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Archaeal DNA-binding protein |
| 21 | d2fbha1 |
|
not modelled |
96.1 |
14 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 22 | c3bj6B_ |
|
not modelled |
96.1 |
13 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulator, marr family;
PDBTitle: crystal structure of marr family transcription regulator sp03579
|
| 23 | c3g3zA_ |
|
not modelled |
95.9 |
14 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator, marr family;
PDBTitle: the structure of nmb1585, a marr family regulator from neisseria2 meningitidis
|
| 24 | d2etha1 |
|
not modelled |
95.9 |
18 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 25 | c3bpxB_ |
|
not modelled |
95.9 |
13 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulator;
PDBTitle: crystal structure of marr
|
| 26 | c3nrvC_ |
|
not modelled |
95.8 |
10 |
PDB header:transcription regulator
Chain: C: PDB Molecule:putative transcriptional regulator (marr/emrr family);
PDBTitle: crystal structure of marr/emrr family transcriptional regulator from2 acinetobacter sp. adp1
|
| 27 | c3e6mD_ |
|
not modelled |
95.7 |
14 |
PDB header:transcription regulator
Chain: D: PDB Molecule:marr family transcriptional regulator;
PDBTitle: the crystal structure of a marr family transcriptional2 regulator from silicibacter pomeroyi dss.
|
| 28 | c2nnnB_ |
|
not modelled |
95.7 |
17 |
PDB header:transcription
Chain: B: PDB Molecule:probable transcriptional regulator;
PDBTitle: crystal structure of probable transcriptional regulator from2 pseudomonas aeruginosa
|
| 29 | c2fa5B_ |
|
not modelled |
95.7 |
10 |
PDB header:transcription
Chain: B: PDB Molecule:transcriptional regulator marr/emrr family;
PDBTitle: the crystal structure of an unliganded multiple antibiotic-2 resistance repressor (marr) from xanthomonas campestris
|
| 30 | d2fxaa1 |
|
not modelled |
95.7 |
24 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 31 | c2rdpA_ |
|
not modelled |
95.6 |
13 |
PDB header:transcription
Chain: A: PDB Molecule:putative transcriptional regulator marr;
PDBTitle: the structure of a marr family protein from bacillus2 stearothermophilus
|
| 32 | d1qwga_ |
|
not modelled |
95.6 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:(2r)-phospho-3-sulfolactate synthase ComA
Family:(2r)-phospho-3-sulfolactate synthase ComA |
| 33 | d1jgsa_ |
|
not modelled |
95.5 |
13 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 34 | c3bjaA_ |
|
not modelled |
95.5 |
10 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator, marr family, putative;
PDBTitle: crystal structure of putative marr-like transcription regulator2 (np_978771.1) from bacillus cereus atcc 10987 at 2.38 a resolution
|
| 35 | d1lnwa_ |
|
not modelled |
95.5 |
9 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 36 | c3k0lA_ |
|
not modelled |
95.4 |
13 |
PDB header:transcription regulator
Chain: A: PDB Molecule:repressor protein;
PDBTitle: crystal structure of putative marr family transcriptional2 regulator from acinetobacter sp. adp
|
| 37 | c2nyxB_ |
|
not modelled |
95.4 |
9 |
PDB header:transcription
Chain: B: PDB Molecule:probable transcriptional regulatory protein, rv1404;
PDBTitle: crystal structure of rv1404 from mycobacterium tuberculosis
|
| 38 | d2a61a1 |
|
not modelled |
95.3 |
20 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 39 | d2fbia1 |
|
not modelled |
95.3 |
12 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 40 | d1lj9a_ |
|
not modelled |
95.3 |
12 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 41 | c2fxaB_ |
|
not modelled |
95.0 |
25 |
PDB header:transcription
Chain: B: PDB Molecule:protease production regulatory protein hpr;
PDBTitle: structure of the protease production regulatory protein hpr from2 bacillus subtilis.
|
| 42 | d1s3ja_ |
|
not modelled |
95.0 |
10 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 43 | d2bv6a1 |
|
not modelled |
95.0 |
10 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 44 | c2pexA_ |
|
not modelled |
95.0 |
18 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator ohrr;
PDBTitle: structure of reduced c22s ohrr from xanthamonas campestris
|
| 45 | c2gxgA_ |
|
not modelled |
95.0 |
18 |
PDB header:transcription
Chain: A: PDB Molecule:146aa long hypothetical transcriptional regulator;
PDBTitle: crystal structure of emrr homolog from hyperthermophilic archaea2 sulfolobus tokodaii strain7
|
| 46 | c3hruA_ |
|
not modelled |
94.9 |
9 |
PDB header:transcription
Chain: A: PDB Molecule:metalloregulator scar;
PDBTitle: crystal structure of scar with bound zn2+
|
| 47 | c3ctaA_ |
|
not modelled |
94.9 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:riboflavin kinase;
PDBTitle: crystal structure of riboflavin kinase from thermoplasma2 acidophilum
|
| 48 | c1nvmG_ |
|
not modelled |
94.9 |
18 |
PDB header:lyase/oxidoreductase
Chain: G: PDB Molecule:4-hydroxy-2-oxovalerate aldolase;
PDBTitle: crystal structure of a bifunctional aldolase-dehydrogenase :2 sequestering a reactive and volatile intermediate
|
| 49 | c3f3xA_ |
|
not modelled |
94.8 |
21 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator, marr family, putative;
PDBTitle: crystal structure of the transcriptional regulator bldr2 from sulfolobus solfataricus
|
| 50 | d2v7fa1 |
|
not modelled |
94.8 |
24 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Rps19E-like |
| 51 | c2ev5B_ |
|
not modelled |
94.7 |
12 |
PDB header:transcription
Chain: B: PDB Molecule:transcriptional regulator mntr;
PDBTitle: bacillus subtilis manganese transport regulator (mntr)2 bound to calcium
|
| 52 | c2qm3A_ |
|
not modelled |
94.7 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:predicted methyltransferase;
PDBTitle: crystal structure of a predicted methyltransferase from pyrococcus2 furiosus
|
| 53 | d3ctaa1 |
|
not modelled |
94.6 |
16 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 54 | c3df8A_ |
|
not modelled |
94.5 |
15 |
PDB header:transcription
Chain: A: PDB Molecule:possible hxlr family transcriptional factor;
PDBTitle: the crystal structure of a possible hxlr family transcriptional factor2 from thermoplasma volcanium gss1
|
| 55 | d1ub9a_ |
|
not modelled |
94.4 |
15 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 56 | c3cdhB_ |
|
not modelled |
94.4 |
16 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulator, marr family;
PDBTitle: crystal structure of the marr family transcriptional regulator spo14532 from silicibacter pomeroyi dss-3
|
| 57 | c3cjnA_ |
|
not modelled |
94.4 |
14 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator, marr family;
PDBTitle: crystal structure of transcriptional regulator, marr family, from2 silicibacter pomeroyi
|
| 58 | c3bleA_ |
|
not modelled |
94.4 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:citramalate synthase from leptospira interrogans;
PDBTitle: crystal structure of the catalytic domain of licms in2 complexed with malonate
|
| 59 | d2f2ea1 |
|
not modelled |
94.4 |
16 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:HxlR-like |
| 60 | c3ivuB_ |
|
not modelled |
94.3 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:homocitrate synthase, mitochondrial;
PDBTitle: homocitrate synthase lys4 bound to 2-og
|
| 61 | c3kp3B_ |
|
not modelled |
94.3 |
7 |
PDB header:transcription regulator/antibiotic
Chain: B: PDB Molecule:transcriptional regulator tcar;
PDBTitle: staphylococcus epidermidis in complex with ampicillin
|
| 62 | c2ftpA_ |
|
not modelled |
94.1 |
10 |
PDB header:lyase
Chain: A: PDB Molecule:hydroxymethylglutaryl-coa lyase;
PDBTitle: crystal structure of hydroxymethylglutaryl-coa lyase from pseudomonas2 aeruginosa
|
| 63 | c3oopA_ |
|
not modelled |
94.1 |
10 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:lin2960 protein;
PDBTitle: the structure of a protein with unknown function from listeria innocua2 clip11262
|
| 64 | d1h4pa_ |
|
not modelled |
93.9 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 65 | c2cw6B_ |
|
not modelled |
93.8 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:hydroxymethylglutaryl-coa lyase, mitochondrial;
PDBTitle: crystal structure of human hmg-coa lyase: insights into2 catalysis and the molecular basis for3 hydroxymethylglutaric aciduria
|
| 66 | c3iz6S_ |
|
not modelled |
93.7 |
18 |
PDB header:ribosome
Chain: S: PDB Molecule:40s ribosomal protein s19 (s19e);
PDBTitle: localization of the small subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
|
| 67 | c2qwwB_ |
|
not modelled |
93.7 |
8 |
PDB header:transcription
Chain: B: PDB Molecule:transcriptional regulator, marr family;
PDBTitle: crystal structure of multiple antibiotic-resistance repressor (marr)2 (yp_013417.1) from listeria monocytogenes 4b f2365 at 2.07 a3 resolution
|
| 68 | d2hr3a1 |
|
not modelled |
93.7 |
12 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 69 | d1yg2a_ |
|
not modelled |
93.6 |
16 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:PadR-like |
| 70 | c3boqB_ |
|
not modelled |
93.5 |
7 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulator, marr family;
PDBTitle: crystal structure of marr family transcriptional regulator from2 silicibacter pomeroyi
|
| 71 | d3broa1 |
|
not modelled |
93.5 |
7 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 72 | d1p4xa2 |
|
not modelled |
93.5 |
13 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 73 | d1z7ua1 |
|
not modelled |
93.4 |
16 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:HxlR-like |
| 74 | c3s2wB_ |
|
not modelled |
93.3 |
14 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulator, marr family;
PDBTitle: the crystal structure of a marr transcriptional regulator from2 methanosarcina mazei go1
|
| 75 | c4a5mH_ |
|
not modelled |
93.3 |
17 |
PDB header:transcription
Chain: H: PDB Molecule:uncharacterized hth-type transcriptional regulator yybr;
PDBTitle: redox regulator hypr in its oxidized form
|
| 76 | c2xznT_ |
|
not modelled |
93.3 |
15 |
PDB header:ribosome
Chain: T: PDB Molecule:rps19e;
PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 2
|
| 77 | c3nqoB_ |
|
not modelled |
93.0 |
14 |
PDB header:transcription
Chain: B: PDB Molecule:marr-family transcriptional regulator;
PDBTitle: crystal structure of a marr family transcriptional regulator (cd1569)2 from clostridium difficile 630 at 2.20 a resolution
|
| 78 | c3hrmA_ |
|
not modelled |
93.0 |
16 |
PDB header:transcription regulator
Chain: A: PDB Molecule:hth-type transcriptional regulator sarz;
PDBTitle: crystal structure of staphylococcus aureus protein sarz in sulfenic2 acid form
|
| 79 | c1p4xA_ |
|
not modelled |
92.5 |
14 |
PDB header:transcription
Chain: A: PDB Molecule:staphylococcal accessory regulator a homologue;
PDBTitle: crystal structure of sars protein from staphylococcus aureus
|
| 80 | d1ad1a_ |
|
not modelled |
92.2 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 81 | d2frha1 |
|
not modelled |
92.1 |
9 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 82 | c3deuB_ |
|
not modelled |
91.8 |
12 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulator slya;
PDBTitle: crystal structure of transcription regulatory protein slya2 from salmonella typhimurium in complex with salicylate3 ligands
|
| 83 | c3eegB_ |
|
not modelled |
91.6 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:2-isopropylmalate synthase;
PDBTitle: crystal structure of a 2-isopropylmalate synthase from2 cytophaga hutchinsonii
|
| 84 | d1hsja1 |
|
not modelled |
91.4 |
14 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 85 | d1z91a1 |
|
not modelled |
91.3 |
20 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 86 | d2fswa1 |
|
not modelled |
91.1 |
14 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:HxlR-like |
| 87 | d1nvma2 |
|
not modelled |
91.0 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:HMGL-like |
| 88 | d1p4xa1 |
|
not modelled |
90.7 |
14 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 89 | c2ou4C_ |
|
not modelled |
90.4 |
13 |
PDB header:isomerase
Chain: C: PDB Molecule:d-tagatose 3-epimerase;
PDBTitle: crystal structure of d-tagatose 3-epimerase from2 pseudomonas cichorii
|
| 90 | d1tw3a1 |
|
not modelled |
90.3 |
20 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Plant O-methyltransferase, N-terminal domain |
| 91 | c3l7wA_ |
|
not modelled |
90.2 |
19 |
PDB header:transcription
Chain: A: PDB Molecule:putative uncharacterized protein smu.1704;
PDBTitle: the crystal structure of smu.1704 from streptococcus mutans ua159
|
| 92 | c1dpuA_ |
|
not modelled |
90.2 |
11 |
PDB header:dna binding protein
Chain: A: PDB Molecule:replication protein a (rpa32) c-terminal domain;
PDBTitle: solution structure of the c-terminal domain of human rpa322 complexed with ung2(73-88)
|
| 93 | d1dpua_ |
|
not modelled |
90.2 |
11 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:C-terminal domain of RPA32 |
| 94 | d2fbka1 |
|
not modelled |
89.8 |
13 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 95 | c3ewbX_ |
|
not modelled |
89.8 |
9 |
PDB header:transferase
Chain: X: PDB Molecule:2-isopropylmalate synthase;
PDBTitle: crystal structure of n-terminal domain of putative 2-2 isopropylmalate synthase from listeria monocytogenes
|
| 96 | d2pb1a1 |
|
not modelled |
89.7 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 97 | c3ecoB_ |
|
not modelled |
89.6 |
6 |
PDB header:transcription
Chain: B: PDB Molecule:mepr;
PDBTitle: crystal structure of mepr, a transcription regulator of the2 staphylococcus aureus multidrug efflux pump mepa
|
| 98 | c3jyvT_ |
|
not modelled |
89.5 |
17 |
PDB header:ribosome
Chain: T: PDB Molecule:s19e protein;
PDBTitle: structure of the 40s rrna and proteins and p/e trna for eukaryotic2 ribosome based on cryo-em map of thermomyces lanuginosus ribosome at3 8.9a resolution
|
| 99 | d1mkma1 |
|
not modelled |
88.9 |
9 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Transcriptional regulator IclR, N-terminal domain |
| 100 | c3fm5D_ |
|
not modelled |
88.5 |
19 |
PDB header:transcription regulator
Chain: D: PDB Molecule:transcriptional regulator;
PDBTitle: x-ray crystal structure of transcriptional regulator (marr family)2 from rhodococcus sp. rha1
|
| 101 | c3navB_ |
|
not modelled |
88.2 |
10 |
PDB header:lyase
Chain: B: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of an alpha subunit of tryptophan synthase from2 vibrio cholerae o1 biovar el tor str. n16961
|
| 102 | c3dp7B_ |
|
not modelled |
88.0 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:sam-dependent methyltransferase;
PDBTitle: crystal structure of sam-dependent methyltransferase from bacteroides2 vulgatus atcc 8482
|
| 103 | c1ydnA_ |
|
not modelled |
87.5 |
8 |
PDB header:lyase
Chain: A: PDB Molecule:hydroxymethylglutaryl-coa lyase;
PDBTitle: crystal structure of the hmg-coa lyase from brucella melitensis,2 northeast structural genomics target lr35.
|
| 104 | c3dx5A_ |
|
not modelled |
87.5 |
8 |
PDB header:lyase
Chain: A: PDB Molecule:uncharacterized protein asbf;
PDBTitle: crystal structure of the probable 3-dhs dehydratase asbf involved in2 the petrobactin synthesis from bacillus anthracis
|
| 105 | d1i60a_ |
|
not modelled |
87.0 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:IolI-like |
| 106 | d1qzza1 |
|
not modelled |
86.7 |
14 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Plant O-methyltransferase, N-terminal domain |
| 107 | d3deua1 |
|
not modelled |
86.7 |
12 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 108 | d1qopa_ |
|
not modelled |
86.7 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 109 | d1hjsa_ |
|
not modelled |
86.4 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 110 | d1gcya2 |
|
not modelled |
86.2 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 111 | c2qw5B_ |
|
not modelled |
86.1 |
14 |
PDB header:isomerase
Chain: B: PDB Molecule:xylose isomerase-like tim barrel;
PDBTitle: crystal structure of a putative sugar phosphate isomerase/epimerase2 (ava4194) from anabaena variabilis atcc 29413 at 1.78 a resolution
|
| 112 | c1xmaA_ |
|
not modelled |
86.1 |
13 |
PDB header:transcription
Chain: A: PDB Molecule:predicted transcriptional regulator;
PDBTitle: structure of a transcriptional regulator from clostridium thermocellum2 cth-833
|
| 113 | d1xmaa_ |
|
not modelled |
86.1 |
13 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:PadR-like |
| 114 | c3mczB_ |
|
not modelled |
86.0 |
23 |
PDB header:transferase
Chain: B: PDB Molecule:o-methyltransferase;
PDBTitle: the structure of an o-methyltransferase family protein from2 burkholderia thailandensis.
|
| 115 | c1ydoC_ |
|
not modelled |
85.5 |
11 |
PDB header:lyase
Chain: C: PDB Molecule:hmg-coa lyase;
PDBTitle: crystal structure of the bacillis subtilis hmg-coa lyase, northeast2 structural genomics target sr181.
|
| 116 | c2dh3A_ |
|
not modelled |
85.5 |
12 |
PDB header:transport protein, signaling protein
Chain: A: PDB Molecule:4f2 cell-surface antigen heavy chain;
PDBTitle: crystal structure of human ed-4f2hc
|
| 117 | c2bmbA_ |
|
not modelled |
85.1 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:folic acid synthesis protein fol1;
PDBTitle: x-ray structure of the bifunctional 6-hydroxymethyl-7,8-2 dihydroxypterin pyrophosphokinase dihydropteroate synthase3 from saccharomyces cerevisiae
|
| 118 | d2q02a1 |
|
not modelled |
84.8 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:IolI-like |
| 119 | d2hzta1 |
|
not modelled |
84.7 |
17 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:HxlR-like |
| 120 | c3aicC_ |
|
not modelled |
84.6 |
21 |
PDB header:transferase
Chain: C: PDB Molecule:glucosyltransferase-si;
PDBTitle: crystal structure of glucansucrase from streptococcus mutans
|