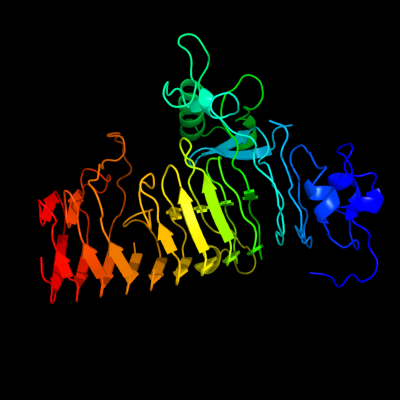
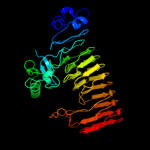
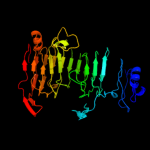
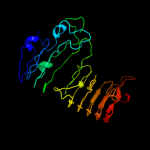
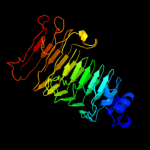
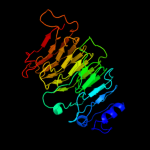
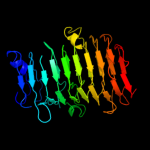
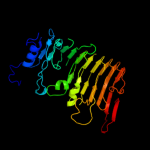
1 c3jurA_
98.5
14
PDB header: hydrolaseChain: A: PDB Molecule: exo-poly-alpha-d-galacturonosidase;PDBTitle: the crystal structure of a hyperthermoactive exopolygalacturonase from2 thermotoga maritima
2 c2uveA_
98.1
21
PDB header: hydrolaseChain: A: PDB Molecule: exopolygalacturonase;PDBTitle: structure of yersinia enterocolitica family 282 exopolygalacturonase
3 d1bhea_
97.8
17
Fold: Single-stranded right-handed beta-helixSuperfamily: Pectin lyase-likeFamily: Galacturonase4 d1rmga_
97.1
16
Fold: Single-stranded right-handed beta-helixSuperfamily: Pectin lyase-likeFamily: Galacturonase5 c3gqnA_
96.5
19
PDB header: viral proteinChain: A: PDB Molecule: preneck appendage protein;PDBTitle: crystal structure of the pre-mature bacteriophage phi29 gene product2 12
6 d1k5ca_
96.3
17
Fold: Single-stranded right-handed beta-helixSuperfamily: Pectin lyase-likeFamily: Galacturonase7 c3eqnB_
95.9
22
PDB header: hydrolaseChain: B: PDB Molecule: glucan 1,3-beta-glucosidase;PDBTitle: crystal structure of beta-1,3-glucanase from phanerochaete2 chrysosporium (lam55a)
8 c2iq7D_
95.8
18
PDB header: hydrolaseChain: D: PDB Molecule: endopolygalacturonase;PDBTitle: crystal structure of the polygalacturonase from colletotrichum lupini2 and its implications for the interaction with polygalacturonase-3 inhibiting proteins
9 c1czfB_
95.6
21
PDB header: hydrolaseChain: B: PDB Molecule: polygalacturonase ii;PDBTitle: endo-polygalacturonase ii from aspergillus niger
10 d1czfa_
95.6
21
Fold: Single-stranded right-handed beta-helixSuperfamily: Pectin lyase-likeFamily: Galacturonase11 c3gq8A_
95.6
24
PDB header: viral proteinChain: A: PDB Molecule: preneck appendage protein;PDBTitle: crystal structure of the bacteriophage phi29 gene product2 12 n-terminal fragment in complex with 2-(n-3 cyclohexylamino)ethane sulfonic acid (ches)
12 d1ia5a_
95.5
21
Fold: Single-stranded right-handed beta-helixSuperfamily: Pectin lyase-likeFamily: Galacturonase13 c2z8gB_
91.7
21
PDB header: hydrolaseChain: B: PDB Molecule: isopullulanase;PDBTitle: aspergillus niger atcc9642 isopullulanase complexed with isopanose
14 d1nhca_
81.0
18
Fold: Single-stranded right-handed beta-helixSuperfamily: Pectin lyase-likeFamily: Galacturonase15 d1hg8a_
79.0
21
Fold: Single-stranded right-handed beta-helixSuperfamily: Pectin lyase-likeFamily: Galacturonase16 d1ogmx2
71.6
19
Fold: Single-stranded right-handed beta-helixSuperfamily: Pectin lyase-likeFamily: Dextranase, catalytic domain17 d1hf2a1
55.1
41
Fold: Single-stranded right-handed beta-helixSuperfamily: Cell-division inhibitor MinC, C-terminal domainFamily: Cell-division inhibitor MinC, C-terminal domain18 c1hf2A_
53.4
41
PDB header: cell division proteinChain: A: PDB Molecule: septum site-determining protein minc;PDBTitle: crystal structure of the bacterial cell-division inhibitor2 minc from t. maritima
19 c1dbgA_
52.2
14
PDB header: lyaseChain: A: PDB Molecule: chondroitinase b;PDBTitle: crystal structure of chondroitinase b
20 d1o88a_
51.5
17
Fold: Single-stranded right-handed beta-helixSuperfamily: Pectin lyase-likeFamily: Pectate lyase-like21 d1pcla_
not modelled
46.7
12
Fold: Single-stranded right-handed beta-helixSuperfamily: Pectin lyase-likeFamily: Pectate lyase-like22 d1xqma_
not modelled
46.5
30
Fold: GFP-likeSuperfamily: GFP-likeFamily: Fluorescent proteins23 c1ogoX_
not modelled
44.9
20
PDB header: hydrolaseChain: X: PDB Molecule: dextranase;PDBTitle: dex49a from penicillium minioluteum complex with isomaltose
24 c3mezA_
not modelled
44.5
20
PDB header: sugar binding proteinChain: A: PDB Molecule: mannose-specific lectin 3 chain 1;PDBTitle: x-ray structural analysis of a mannose specific lectin from dutch2 crocus (crocus vernus)
25 c2xvsA_
not modelled
43.6
19
PDB header: antitumor proteinChain: A: PDB Molecule: tetratricopeptide repeat protein 5;PDBTitle: crystal structure of human ttc5 (strap) c-terminal ob2 domain
26 d2f2ha1
not modelled
38.8
28
Fold: Putative glucosidase YicI, C-terminal domainSuperfamily: Putative glucosidase YicI, C-terminal domainFamily: Putative glucosidase YicI, C-terminal domain27 d1bn8a_
not modelled
38.6
22
Fold: Single-stranded right-handed beta-helixSuperfamily: Pectin lyase-likeFamily: Pectate lyase-like28 c2pyhB_
not modelled
37.8
19
PDB header: isomeraseChain: B: PDB Molecule: poly(beta-d-mannuronate) c5 epimerase 4;PDBTitle: azotobacter vinelandii mannuronan c-5 epimerase alge4 a-module2 complexed with mannuronan trisaccharide
29 d1j3pa_
not modelled
33.3
22
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Glucose-6-phosphate isomerase, GPI30 c1nkgA_
not modelled
30.2
30
PDB header: lyaseChain: A: PDB Molecule: rhamnogalacturonase b;PDBTitle: rhamnogalacturonan lyase from aspergillus aculeatus
31 d1idka_
not modelled
29.8
21
Fold: Single-stranded right-handed beta-helixSuperfamily: Pectin lyase-likeFamily: Pectin lyase32 c2dd9C_
not modelled
29.2
20
PDB header: luminescent proteinChain: C: PDB Molecule: green fluorescent protein;PDBTitle: a mutant of gfp-like protein from chiridius poppei
33 d2qqra1
not modelled
28.6
37
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: Tudor domain34 c1vblA_
not modelled
26.2
19
PDB header: lyaseChain: A: PDB Molecule: pectate lyase 47;PDBTitle: structure of the thermostable pectate lyase pl 47
35 c2qy1B_
not modelled
26.1
14
PDB header: lyaseChain: B: PDB Molecule: pectate lyase ii;PDBTitle: pectate lyase a31g/r236f from xanthomonas campestris
36 c2dpfB_
not modelled
25.6
25
PDB header: plant proteinChain: B: PDB Molecule: curculin;PDBTitle: crystal structure of curculin1 homodimer
37 c3rrrB_
not modelled
25.3
32
PDB header: viral proteinChain: B: PDB Molecule: fusion glycoprotein f0;PDBTitle: structure of the rsv f protein in the post-fusion conformation
38 d1bwud_
not modelled
25.0
13
Fold: beta-Prism IISuperfamily: alpha-D-mannose-specific plant lectinsFamily: alpha-D-mannose-specific plant lectins39 c3jyvT_
not modelled
24.8
23
PDB header: ribosomeChain: T: PDB Molecule: s19e protein;PDBTitle: structure of the 40s rrna and proteins and p/e trna for eukaryotic2 ribosome based on cryo-em map of thermomyces lanuginosus ribosome at3 8.9a resolution
40 d1x82a_
not modelled
24.7
23
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Glucose-6-phosphate isomerase, GPI41 c3r0eC_
not modelled
23.3
33
PDB header: sugar binding proteinChain: C: PDB Molecule: lectin;PDBTitle: structure of remusatia vivipara lectin
42 d1s0ua2
not modelled
21.3
78
Fold: Elongation factor/aminomethyltransferase common domainSuperfamily: EF-Tu/eEF-1alpha/eIF2-gamma C-terminal domainFamily: EF-Tu/eEF-1alpha/eIF2-gamma C-terminal domain43 c3rkiB_
not modelled
21.0
32
PDB header: viral proteinChain: B: PDB Molecule: fusion glycoprotein f0;PDBTitle: structural basis for immunization with post-fusion rsv f to elicit2 high neutralizing antibody titers
44 c3u5ga_
not modelled
19.6
33
PDB header: ribosomeChain: A: PDB Molecule: 40s ribosomal protein s0-a;PDBTitle: the structure of the eukaryotic ribosome at 3.0 a resolution
45 c3cfhB_
not modelled
19.5
31
PDB header: fluorescent proteinChain: B: PDB Molecule: gfp-like photoswitchable fluorescent protein;PDBTitle: photoswitchable red fluorescent protein psrfp, off-state
46 d1jpca_
not modelled
19.5
23
Fold: beta-Prism IISuperfamily: alpha-D-mannose-specific plant lectinsFamily: alpha-D-mannose-specific plant lectins47 d1npla_
not modelled
19.0
28
Fold: beta-Prism IISuperfamily: alpha-D-mannose-specific plant lectinsFamily: alpha-D-mannose-specific plant lectins48 d1pxza_
not modelled
18.7
12
Fold: Single-stranded right-handed beta-helixSuperfamily: Pectin lyase-likeFamily: Pectate lyase-like49 c2xzn5_
not modelled
17.9
28
PDB header: ribosomeChain: 5: PDB Molecule: ribosomal protein s26e containing protein;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 2
50 c3lasA_
not modelled
17.2
31
PDB header: lyaseChain: A: PDB Molecule: putative carbonic anhydrase;PDBTitle: crystal structure of carbonic anhydrase from streptococcus mutans to2 1.4 angstrom resolution
51 c3nezB_
not modelled
16.8
23
PDB header: fluorescent proteinChain: B: PDB Molecule: mrojoa;PDBTitle: mrojoa
52 c3kv4A_
not modelled
16.6
42
PDB header: h3k4me3 binding protein, transferaseChain: A: PDB Molecule: phd finger protein 8;PDBTitle: structure of phf8 in complex with histone h3
53 c1z8rA_
not modelled
15.8
50
PDB header: hydrolaseChain: A: PDB Molecule: coxsackievirus b4 polyprotein;PDBTitle: 2a cysteine proteinase from human coxsackievirus b4 (strain2 jvb / benschoten / new york / 51)
54 d1xd5a_
not modelled
15.7
25
Fold: beta-Prism IISuperfamily: alpha-D-mannose-specific plant lectinsFamily: alpha-D-mannose-specific plant lectins55 c3iz6S_
not modelled
15.6
16
PDB header: ribosomeChain: S: PDB Molecule: 40s ribosomal protein s19 (s19e);PDBTitle: localization of the small subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
56 c3mezB_
not modelled
15.1
10
PDB header: sugar binding proteinChain: B: PDB Molecule: mannose-specific lectin 3 chain 2;PDBTitle: x-ray structural analysis of a mannose specific lectin from dutch2 crocus (crocus vernus)
57 d1o12a1
not modelled
15.1
17
Fold: Composite domain of metallo-dependent hydrolasesSuperfamily: Composite domain of metallo-dependent hydrolasesFamily: N-acetylglucosamine-6-phosphate deacetylase, NagA58 d2hv2a1
not modelled
14.6
19
Fold: SCP-likeSuperfamily: SCP-likeFamily: EF1021 C-terminal domain-like59 d2hrva_
not modelled
14.4
33
Fold: Trypsin-like serine proteasesSuperfamily: Trypsin-like serine proteasesFamily: Viral cysteine protease of trypsin fold60 c2nlwA_
not modelled
14.4
16
PDB header: translationChain: A: PDB Molecule: eukaryotic translation initiation factor 3PDBTitle: solution structure of the rrm domain of human eukaryotic2 initiation factor 3b
61 d2c5lc1
not modelled
13.9
43
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: Ras-binding domain, RBD62 c2wulB_
not modelled
13.9
27
PDB header: oxidoreductaseChain: B: PDB Molecule: glutaredoxin related protein 5;PDBTitle: crystal structure of the human glutaredoxin 5 with bound2 glutathione in an fes cluster
63 c3a0cA_
not modelled
13.8
19
PDB header: sugar binding proteinChain: A: PDB Molecule: mannose/sialic acid-binding lectin;PDBTitle: crystal structure of an anti-hiv mannose-binding lectin from2 polygonatum cyrtonema hua
64 c3grhA_
not modelled
13.6
17
PDB header: hydrolaseChain: A: PDB Molecule: acyl-coa thioester hydrolase ybgc;PDBTitle: crystal structure of escherichia coli ybhc
65 d2v7fa1
not modelled
13.2
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Rps19E-like66 c3b08H_
not modelled
13.1
43
PDB header: signaling protein/metal binding proteinChain: H: PDB Molecule: ranbp-type and c3hc4-type zinc finger-containing protein 1;PDBTitle: crystal structure of the mouse hoil1-l-nzf in complex with linear di-2 ubiquitin
67 c3ipzA_
not modelled
12.9
40
PDB header: electron transport, oxidoreductaseChain: A: PDB Molecule: monothiol glutaredoxin-s14, chloroplastic;PDBTitle: crystal structure of arabidopsis monothiol glutaredoxin atgrxcp
68 c2e7pC_
not modelled
12.7
47
PDB header: electron transportChain: C: PDB Molecule: glutaredoxin;PDBTitle: crystal structure of the holo form of glutaredoxin c1 from populus2 tremula x tremuloides
69 d1jnya2
not modelled
12.6
19
Fold: Elongation factor/aminomethyltransferase common domainSuperfamily: EF-Tu/eEF-1alpha/eIF2-gamma C-terminal domainFamily: EF-Tu/eEF-1alpha/eIF2-gamma C-terminal domain70 c3hz7A_
not modelled
12.5
50
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of the sira-like protein (dsy4693) from2 desulfitobacterium hafniense, northeast structural genomics3 consortium target dhr2a
71 c2z6zA_
not modelled
12.4
26
PDB header: fluorescent proteinChain: A: PDB Molecule: fluorescent protein dronpa;PDBTitle: crystal structure of a photoswitchable gfp-like protein2 dronpa in the bright-state
72 c3l4nA_
not modelled
12.4
27
PDB header: oxidoreductaseChain: A: PDB Molecule: monothiol glutaredoxin-6;PDBTitle: crystal structure of yeast monothiol glutaredoxin grx6
73 c2jacA_
not modelled
11.7
47
PDB header: electron transportChain: A: PDB Molecule: glutaredoxin-1;PDBTitle: glutaredoxin grx1p c30s mutant from yeast
74 c3c1sA_
not modelled
10.9
50
PDB header: oxidoreductaseChain: A: PDB Molecule: glutaredoxin-1;PDBTitle: crystal structure of grx1 in glutathionylated form
75 d1moua_
not modelled
10.9
23
Fold: GFP-likeSuperfamily: GFP-likeFamily: Fluorescent proteins76 d1jhba_
not modelled
10.7
27
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Thioltransferase77 d1kj1d_
not modelled
10.6
14
Fold: beta-Prism IISuperfamily: alpha-D-mannose-specific plant lectinsFamily: alpha-D-mannose-specific plant lectins78 c3r0eD_
not modelled
10.6
20
PDB header: sugar binding proteinChain: D: PDB Molecule: lectin;PDBTitle: structure of remusatia vivipara lectin
79 d1nkga3
not modelled
10.1
20
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: Rhamnogalacturonase B, RhgB, N-terminal domain80 c3pu3A_
not modelled
9.9
45
PDB header: protein bindingChain: A: PDB Molecule: phd finger protein 2;PDBTitle: phf2 jumonji domain-nog complex
81 c3k3nA_
not modelled
9.8
50
PDB header: oxidoreductaseChain: A: PDB Molecule: phd finger protein 8;PDBTitle: crystal structure of the catalytic core domain of human phf8
82 d1ggxa_
not modelled
9.8
27
Fold: GFP-likeSuperfamily: GFP-likeFamily: Fluorescent proteins83 c2gw4D_
not modelled
9.8
23
PDB header: luminescent proteinChain: D: PDB Molecule: kaede;PDBTitle: crystal structure of stony coral fluorescent protein kaede, red form
84 c2w3nA_
not modelled
9.7
31
PDB header: lyaseChain: A: PDB Molecule: carbonic anhydrase 2;PDBTitle: structure and inhibition of the co2-sensing carbonic2 anhydrase can2 from the pathogenic fungus cryptococcus3 neoformans
85 d1ktea_
not modelled
9.7
27
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Thioltransferase86 d2rkya1
not modelled
9.7
57
Fold: FnI-like domainSuperfamily: FnI-like domainFamily: Fibronectin type I module87 d1bwua_
not modelled
9.6
17
Fold: beta-Prism IISuperfamily: alpha-D-mannose-specific plant lectinsFamily: alpha-D-mannose-specific plant lectins88 d1jdqa_
not modelled
9.4
50
Fold: IF3-likeSuperfamily: SirA-likeFamily: SirA-like89 c3gztF_
not modelled
9.3
86
PDB header: virusChain: F: PDB Molecule: outer capsid glycoprotein vp7;PDBTitle: vp7 recoated rotavirus dlp
90 d2naca2
not modelled
9.3
20
Fold: Flavodoxin-likeSuperfamily: Formate/glycerate dehydrogenase catalytic domain-likeFamily: Formate/glycerate dehydrogenases, substrate-binding domain91 d1lbqa_
not modelled
9.3
29
Fold: Chelatase-likeSuperfamily: ChelataseFamily: Ferrochelatase92 d1xd6a_
not modelled
9.2
30
Fold: beta-Prism IISuperfamily: alpha-D-mannose-specific plant lectinsFamily: alpha-D-mannose-specific plant lectins93 c1ylkA_
not modelled
9.2
25
PDB header: unknown functionChain: A: PDB Molecule: hypothetical protein rv1284/mt1322;PDBTitle: crystal structure of rv1284 from mycobacterium tuberculosis in complex2 with thiocyanate
94 d2cg7a1
not modelled
9.1
57
Fold: FnI-like domainSuperfamily: FnI-like domainFamily: Fibronectin type I module95 c2zo7A_
not modelled
9.0
24
PDB header: luminescent proteinChain: A: PDB Molecule: cyan/green-emitting gfp-like protein, kusabira-cyan mutantPDBTitle: crystal structure of a kusabira-cyan mutant (kcy-r1), a cyan/green-2 emitting gfp-like protein
96 c2hzfA_
not modelled
8.9
27
PDB header: electron transport, oxidoreductaseChain: A: PDB Molecule: glutaredoxin-1;PDBTitle: crystal structures of a poxviral glutaredoxin in the oxidized and2 reduced states show redox-correlated structural changes
97 d2rkya2
not modelled
8.9
71
Fold: FnI-like domainSuperfamily: FnI-like domainFamily: Fibronectin type I module98 c2eceA_
not modelled
8.7
25
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: 462aa long hypothetical selenium-binding protein;PDBTitle: x-ray structure of hypothetical selenium-binding protein2 from sulfolobus tokodaii, st0059
99 c2ddcA_
not modelled
8.6
34
PDB header: luminescent proteinChain: A: PDB Molecule: photoconvertible fluorescent protein;PDBTitle: unique behavior of a histidine responsible for an2 engineered green-to-red photoconversion process