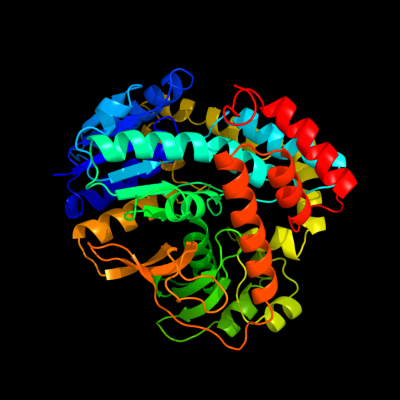
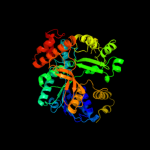
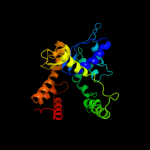
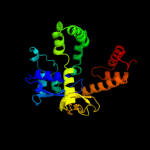
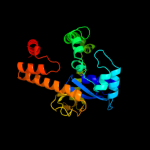
1 c1u8xX_
100.0
25
PDB header: hydrolaseChain: X: PDB Molecule: maltose-6'-phosphate glucosidase;PDBTitle: crystal structure of glva from bacillus subtilis, a metal-requiring,2 nad-dependent 6-phospho-alpha-glucosidase
2 c1s6yA_
100.0
53
PDB header: hydrolaseChain: A: PDB Molecule: 6-phospho-beta-glucosidase;PDBTitle: 2.3a crystal structure of phospho-beta-glucosidase
3 c1up6F_
100.0
32
PDB header: hydrolaseChain: F: PDB Molecule: 6-phospho-beta-glucosidase;PDBTitle: structure of the 6-phospho-beta glucosidase from thermotoga2 maritima at 2.55 angstrom resolution in the tetragonal form3 with manganese, nad+ and glucose-6-phosphate
4 c1vjtA_
100.0
19
PDB header: hydrolaseChain: A: PDB Molecule: alpha-glucosidase;PDBTitle: crystal structure of alpha-glucosidase (tm0752) from thermotoga2 maritima at 2.50 a resolution
5 c1obbB_
100.0
25
PDB header: hydrolaseChain: B: PDB Molecule: alpha-glucosidase;PDBTitle: alpha-glucosidase a, agla, from thermotoga maritima in2 complex with maltose and nad+
6 c3fefB_
100.0
21
PDB header: hydrolaseChain: B: PDB Molecule: putative glucosidase lpld;PDBTitle: crystal structure of putative glucosidase lpld from2 bacillus subtilis
7 d1u8xx2
100.0
18
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: AglA-like glucosidase8 d1s6ya2
100.0
43
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: AglA-like glucosidase9 d1up7a2
100.0
29
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: AglA-like glucosidase10 d1obba2
100.0
21
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: AglA-like glucosidase11 d1vjta2
100.0
15
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: AglA-like glucosidase12 c3p7mC_
100.0
21
PDB header: oxidoreductaseChain: C: PDB Molecule: malate dehydrogenase;PDBTitle: structure of putative lactate dehydrogenase from francisella2 tularensis subsp. tularensis schu s4
13 c1pzfD_
100.0
17
PDB header: oxidoreductaseChain: D: PDB Molecule: lactate dehydrogenase;PDBTitle: t.gondii ldh1 ternary complex with apad+ and oxalate
14 c2ldxA_
100.0
18
PDB header: oxidoreductase(choh(d)-nad(a))Chain: A: PDB Molecule: apo-lactate dehydrogenase;PDBTitle: characterization of the antigenic sites on the refined 3-2 angstroms resolution structure of mouse testicular lactate3 dehydrogenase c4
15 c2v65A_
100.0
18
PDB header: oxidoreductaseChain: A: PDB Molecule: l-lactate dehydrogenase a chain;PDBTitle: apo ldh from the psychrophile c. gunnari
16 d1vjta1
100.0
26
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like17 c1ez4B_
100.0
19
PDB header: oxidoreductaseChain: B: PDB Molecule: lactate dehydrogenase;PDBTitle: crystal structure of non-allosteric l-lactate dehydrogenase2 from lactobacillus pentosus at 2.3 angstrom resolution
18 c2fnzA_
100.0
19
PDB header: oxidoreductaseChain: A: PDB Molecule: lactate dehydrogenase;PDBTitle: crystal structure of the lactate dehydrogenase from cryptosporidium2 parvum complexed with cofactor (b-nicotinamide adenine dinucleotide)3 and inhibitor (oxamic acid)
19 c2hjrK_
100.0
20
PDB header: oxidoreductaseChain: K: PDB Molecule: malate dehydrogenase;PDBTitle: crystal structure of cryptosporidium parvum malate2 dehydrogenase
20 d1s6ya1
100.0
68
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like21 c8ldhA_
not modelled
100.0
15
PDB header: oxidoreductase(choh(d)-nad(a))Chain: A: PDB Molecule: m4 apo-lactate dehydrogenase;PDBTitle: refined crystal structure of dogfish m4 apo-lactate2 dehydrogenase
22 c3d0oA_
not modelled
100.0
18
PDB header: oxidoreductaseChain: A: PDB Molecule: l-lactate dehydrogenase 1;PDBTitle: crystal structure of lactate dehydrogenase from2 staphylococcus aureus
23 c3gviB_
not modelled
100.0
18
PDB header: oxidoreductaseChain: B: PDB Molecule: malate dehydrogenase;PDBTitle: crystal structure of lactate/malate dehydrogenase from2 brucella melitensis in complex with adp
24 c1a5zA_
not modelled
100.0
18
PDB header: oxidoreductaseChain: A: PDB Molecule: l-lactate dehydrogenase;PDBTitle: lactate dehydrogenase from thermotoga maritima (tmldh)
25 c1lldA_
not modelled
100.0
18
PDB header: oxidoreductase(choh (d)-nad (a))Chain: A: PDB Molecule: l-lactate dehydrogenase;PDBTitle: molecular basis of allosteric activation of bacterial l-lactate2 dehydrogenase
26 d1u8xx1
not modelled
100.0
35
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like27 c3tl2A_
not modelled
100.0
18
PDB header: oxidoreductaseChain: A: PDB Molecule: malate dehydrogenase;PDBTitle: crystal structure of bacillus anthracis str. ames malate dehydrogenase2 in closed conformation.
28 c1u4sA_
not modelled
100.0
18
PDB header: oxidoreductaseChain: A: PDB Molecule: l-lactate dehydrogenase;PDBTitle: plasmodium falciparum lactate dehydrogenase complexed with 2,6-2 naphthalenedisulphonic acid
29 d1up7a1
not modelled
100.0
38
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like30 c1ojuA_
not modelled
100.0
19
PDB header: oxidoreductaseChain: A: PDB Molecule: malate dehydrogenase;PDBTitle: 2.8 a resolution structure of malate dehydrogenase from2 archaeoglobus fulgidus in complex with etheno-nad.
31 c1llcA_
not modelled
100.0
17
PDB header: oxidoreductase(choh(d)-nad(a))Chain: A: PDB Molecule: l-lactate dehydrogenase;PDBTitle: structure determination of the allosteric l-lactate dehydrogenase from2 lactobacillus casei at 3.0 angstroms resolution
32 c1hyhA_
not modelled
100.0
20
PDB header: oxidoreductase (choh(d)-nad+(a))Chain: A: PDB Molecule: l-2-hydroxyisocaproate dehydrogenase;PDBTitle: crystal structure of l-2-hydroxyisocaproate dehydrogenase from2 lactobacillus confusus at 2.2 angstroms resolution-an example of3 strong asymmetry between subunits
33 c3nepX_
not modelled
100.0
18
PDB header: oxidoreductaseChain: X: PDB Molecule: malate dehydrogenase;PDBTitle: 1.55a resolution structure of malate dehydrogenase from salinibacter2 ruber
34 c3pqeD_
not modelled
100.0
19
PDB header: oxidoreductaseChain: D: PDB Molecule: l-lactate dehydrogenase;PDBTitle: crystal structure of l-lactate dehydrogenase from bacillus subtilis2 with h171c mutation
35 c1ur5C_
not modelled
100.0
20
PDB header: oxidoreductaseChain: C: PDB Molecule: malate dehydrogenase;PDBTitle: stabilization of a tetrameric malate dehydrogenase by2 introduction of a disulfide bridge at the dimer/dimer3 interface
36 c1hygA_
not modelled
100.0
16
PDB header: oxidoreductaseChain: A: PDB Molecule: l-lactate/malate dehydrogenase;PDBTitle: crystal structure of mj0490 gene product, the family of2 lactate/malate dehydrogenase
37 d1obba1
not modelled
100.0
26
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like38 c7mdhA_
not modelled
100.0
16
PDB header: chloroplastic malate dehydrogenaseChain: A: PDB Molecule: protein (malate dehydrogenase);PDBTitle: structural basis for light acitvation of a chloroplast enzyme. the2 structure of sorghum nadp-malate dehydrogenase in its oxidized form
39 c2e37B_
not modelled
100.0
18
PDB header: oxidoreductaseChain: B: PDB Molecule: l-lactate dehydrogenase;PDBTitle: structure of tt0471 protein from thermus thermophilus
40 c1b8vA_
not modelled
100.0
14
PDB header: oxidoreductaseChain: A: PDB Molecule: protein (malate dehydrogenase);PDBTitle: malate dehydrogenase from aquaspirillum arcticum
41 c2v6bB_
not modelled
100.0
19
PDB header: oxidoreductaseChain: B: PDB Molecule: l-lactate dehydrogenase;PDBTitle: crystal structure of lactate dehydrogenase from deinococcus2 radiodurans (apo form)
42 c1gv1D_
not modelled
100.0
22
PDB header: oxidoreductaseChain: D: PDB Molecule: malate dehydrogenase;PDBTitle: structural basis for thermophilic protein stability:2 structures of thermophilic and mesophilic malate3 dehydrogenases
43 c2pwzG_
not modelled
100.0
15
PDB header: oxidoreductaseChain: G: PDB Molecule: malate dehydrogenase;PDBTitle: crystal structure of the apo form of e.coli malate dehydrogenase
44 c2d4aC_
not modelled
100.0
16
PDB header: oxidoreductaseChain: C: PDB Molecule: malate dehydrogenase;PDBTitle: structure of the malate dehydrogenase from aeropyrum pernix
45 c5mdhB_
not modelled
100.0
16
PDB header: oxidoreductaseChain: B: PDB Molecule: malate dehydrogenase;PDBTitle: crystal structure of ternary complex of porcine cytoplasmic malate2 dehydrogenase alpha-ketomalonate and tnad at 2.4 angstroms resolution
46 c1ldbA_
not modelled
100.0
21
PDB header: oxidoreductase(choh(d)-nad(a))Chain: A: PDB Molecule: apo-l-lactate dehydrogenase;PDBTitle: structure determination and refinement of bacillus2 stearothermophilus lactate dehydrogenase
47 c1y6jA_
not modelled
100.0
18
PDB header: oxidoreductaseChain: A: PDB Molecule: l-lactate dehydrogenase;PDBTitle: l-lactate dehydrogenase from clostridium thermocellum cth-1135
48 c1wziA_
not modelled
100.0
16
PDB header: oxidoreductaseChain: A: PDB Molecule: malate dehydrogenase;PDBTitle: structural basis for alteration of cofactor specificity of2 malate dehydrogenase from thermus flavus
49 c3dl2A_
not modelled
100.0
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: ubiquitin-conjugating enzyme e2 variant 3;PDBTitle: hexagonal structure of the ldh domain of human ubiquitin-2 conjugating enzyme e2-like isoform a
50 c3fi9B_
not modelled
100.0
19
PDB header: oxidoreductaseChain: B: PDB Molecule: malate dehydrogenase;PDBTitle: crystal structure of malate dehydrogenase from porphyromonas2 gingivalis
51 c1smkD_
not modelled
100.0
14
PDB header: oxidoreductaseChain: D: PDB Molecule: malate dehydrogenase, glyoxysomal;PDBTitle: mature and translocatable forms of glyoxysomal malate2 dehydrogenase have different activities and stabilities3 but similar crystal structures
52 c1sevA_
not modelled
100.0
14
PDB header: oxidoreductaseChain: A: PDB Molecule: malate dehydrogenase, glyoxysomal precursor;PDBTitle: mature and translocatable forms of glyoxysomal malate2 dehydrogenase have different activities and stabilities3 but similar crystal structures
53 c2dfdD_
not modelled
100.0
15
PDB header: oxidoreductaseChain: D: PDB Molecule: malate dehydrogenase;PDBTitle: crystal structure of human malate dehydrogenase type 2
54 c1mldA_
not modelled
100.0
15
PDB header: oxidoreductase(nad(a)-choh(d))Chain: A: PDB Molecule: malate dehydrogenase;PDBTitle: refined structure of mitochondrial malate dehydrogenase2 from porcine heart and the consensus structure for3 dicarboxylic acid oxidoreductases
55 c2hlpB_
not modelled
100.0
16
PDB header: oxidoreductaseChain: B: PDB Molecule: malate dehydrogenase;PDBTitle: crystal structure of the e267r mutant of a halophilic2 malate dehydrogenase in the apo form
56 d1i0za1
not modelled
99.9
21
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like57 d1o6za1
not modelled
99.9
17
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like58 d1pzga1
not modelled
99.9
17
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like59 d1i10a1
not modelled
99.9
21
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like60 d1t2da1
not modelled
99.9
21
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like61 d5ldha1
not modelled
99.9
22
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like62 d9ldta1
not modelled
99.9
20
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like63 d1y7ta1
not modelled
99.9
19
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like64 d1a5za1
not modelled
99.9
23
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like65 d1ojua1
not modelled
99.9
18
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like66 d1b8pa1
not modelled
99.9
18
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like67 d1ldna1
not modelled
99.9
22
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like68 d1guza1
not modelled
99.9
21
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like69 d2ldxa1
not modelled
99.9
23
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like70 d1llda1
not modelled
99.9
22
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like71 d1ldma1
not modelled
99.9
16
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like72 d2cmda1
not modelled
99.9
22
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like73 d1hyha1
not modelled
99.9
20
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like74 d5mdha1
not modelled
99.9
18
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like75 d1llca1
not modelled
99.9
21
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like76 d7mdha1
not modelled
99.9
16
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like77 d1gv0a1
not modelled
99.9
20
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like78 d1civa1
not modelled
99.8
17
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like79 d1uxja1
not modelled
99.8
19
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like80 d1y6ja1
not modelled
99.8
20
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like81 d1ez4a1
not modelled
99.7
30
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like82 d1hyea1
not modelled
99.7
18
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like83 d1mlda1
not modelled
99.6
20
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like84 d9ldta2
not modelled
99.6
15
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain85 d1i10a2
not modelled
99.6
16
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain86 d1i0za2
not modelled
99.5
16
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain87 d1oc4a2
not modelled
99.5
13
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain88 d1ldma2
not modelled
99.5
15
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain89 d1a5za2
not modelled
99.5
14
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain90 d1t2da2
not modelled
99.5
13
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain91 d2ldxa2
not modelled
99.5
17
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain92 d1pzga2
not modelled
99.5
14
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain93 d1llda2
not modelled
99.5
16
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain94 d5mdha2
not modelled
99.5
16
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain95 d1ldna2
not modelled
99.4
18
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain96 d1civa2
not modelled
99.4
15
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain97 d7mdha2
not modelled
99.4
13
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain98 d1uxja2
not modelled
99.4
21
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain99 d1gv0a2
not modelled
99.4
19
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain100 d1guza2
not modelled
99.4
19
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain101 d1o6za2
not modelled
99.4
15
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain102 d1ez4a2
not modelled
99.4
18
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain103 d1hyha2
not modelled
99.4
18
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain104 d1b8pa2
not modelled
99.4
12
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain105 d1y7ta2
not modelled
99.4
15
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain106 d1hyea2
not modelled
99.3
17
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain107 d1ojua2
not modelled
99.3
19
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain108 d1y6ja2
not modelled
99.3
15
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain109 d1llca2
not modelled
99.2
14
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain110 d1mlda2
not modelled
99.1
11
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain111 d2cmda2
not modelled
99.0
11
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain112 c3prjB_
not modelled
98.8
18
PDB header: oxidoreductaseChain: B: PDB Molecule: udp-glucose 6-dehydrogenase;PDBTitle: role of packing defects in the evolution of allostery and induced fit2 in human udp-glucose dehydrogenase.
113 c1m75B_
not modelled
98.7
18
PDB header: oxidoreductaseChain: B: PDB Molecule: 3-hydroxyacyl-coa dehydrogenase;PDBTitle: crystal structure of the n208s mutant of l-3-hydroxyacyl-2 coa dehydrogenase in complex with nad and acetoacetyl-coa
114 c1mv8A_
not modelled
98.6
21
PDB header: oxidoreductaseChain: A: PDB Molecule: gdp-mannose 6-dehydrogenase;PDBTitle: 1.55 a crystal structure of a ternary complex of gdp-mannose2 dehydrogenase from psuedomonas aeruginosa
115 d1dlja2
not modelled
98.6
21
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain116 d1txga2
not modelled
98.5
10
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain117 c3k96B_
not modelled
98.5
16
PDB header: oxidoreductaseChain: B: PDB Molecule: glycerol-3-phosphate dehydrogenase [nad(p)+];PDBTitle: 2.1 angstrom resolution crystal structure of glycerol-3-phosphate2 dehydrogenase (gpsa) from coxiella burnetii
118 c2y0dB_
not modelled
98.5
17
PDB header: oxidoreductaseChain: B: PDB Molecule: udp-glucose dehydrogenase;PDBTitle: bcec mutation y10k
119 c2q3eH_
not modelled
98.5
16
PDB header: oxidoreductaseChain: H: PDB Molecule: udp-glucose 6-dehydrogenase;PDBTitle: structure of human udp-glucose dehydrogenase complexed with nadh and2 udp-glucose
120 c3plnA_
not modelled
98.5
20
PDB header: oxidoreductaseChain: A: PDB Molecule: udp-glucose 6-dehydrogenase;PDBTitle: crystal structure of klebsiella pneumoniae udp-glucose 6-dehydrogenase2 complexed with udp-glucose