1 c2nvoA_
99.7
14
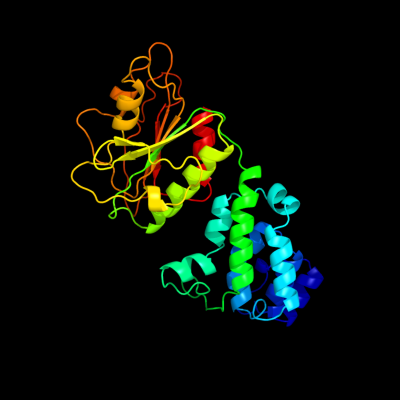
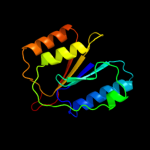
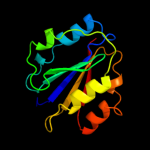
PDB header: rna binding proteinChain: A: PDB Molecule: ro sixty-related protein, rsr;PDBTitle: crystal structure of deinococcus radiodurans ro (rsr) protein
2 c3ibsA_
99.7
16
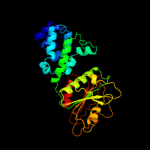
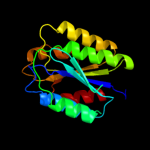
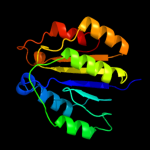
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved hypothetical protein batb;PDBTitle: crystal structure of conserved hypothetical protein batb from2 bacteroides thetaiotaomicron
3 c2x5nA_
99.6
11
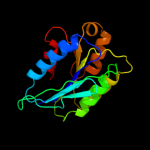
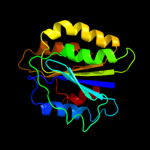
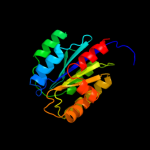
PDB header: nuclear proteinChain: A: PDB Molecule: 26s proteasome regulatory subunit rpn10;PDBTitle: crystal structure of the sprpn10 vwa domain
4 c2i6sA_
99.6
18
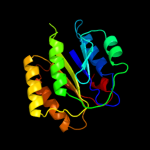
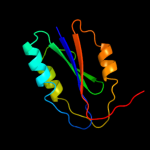
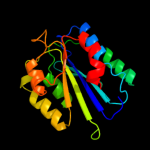
PDB header: hydrolaseChain: A: PDB Molecule: complement c2a fragment;PDBTitle: complement component c2a
5 c1rs0A_
99.6
19
PDB header: hydrolaseChain: A: PDB Molecule: complement factor b;PDBTitle: crystal structure analysis of the bb segment of factor b2 complexed with di-isopropyl-phosphate (dip)
6 c2x31F_
99.6
19
PDB header: ligaseChain: F: PDB Molecule: magnesium-chelatase 60 kda subunit;PDBTitle: modelling of the complex between subunits bchi and bchd of magnesium2 chelatase based on single-particle cryo-em reconstruction at 7.5 ang
7 c2ok5A_
99.6
19
PDB header: hydrolaseChain: A: PDB Molecule: complement factor b;PDBTitle: human complement factor b
8 d1jeyb2
99.5
22
Fold: vWA-likeSuperfamily: vWA-likeFamily: Ku80 subunit N-terminal domain9 d2ok5a1
99.5
19
Fold: vWA-likeSuperfamily: vWA-likeFamily: Integrin A (or I) domain10 d1yvra2
99.5
20
Fold: vWA-likeSuperfamily: vWA-likeFamily: RoRNP C-terminal domain-like11 c1yvrA_
99.5
15
PDB header: rna binding proteinChain: A: PDB Molecule: 60-kda ss-a/ro ribonucleoprotein;PDBTitle: ro autoantigen
12 d1jeya2
99.5
21
Fold: vWA-likeSuperfamily: vWA-likeFamily: Ku70 subunit N-terminal domain13 d1shux_
99.4
18
Fold: vWA-likeSuperfamily: vWA-likeFamily: Integrin A (or I) domain14 c2b2xB_
99.3
15
PDB header: immune systemChain: B: PDB Molecule: integrin alpha-1;PDBTitle: vla1 rdeltah i-domain complexed with a quadruple mutant of the aqc22 fab
15 c2iueA_
99.3
21
PDB header: membrane proteinChain: A: PDB Molecule: pactolus i-domain;PDBTitle: pactolus i-domain: functional switching of the rossmann2 fold
16 c2xggB_
99.3
19
PDB header: hydrolaseChain: B: PDB Molecule: microneme protein 2;PDBTitle: structure of toxoplasma gondii micronemal protein 2 a_i2 domain
17 d1pd0a3
99.3
22
Fold: vWA-likeSuperfamily: vWA-likeFamily: Trunk domain of Sec23/2418 c3n2nC_
99.3
18
PDB header: toxin receptorChain: C: PDB Molecule: anthrax toxin receptor 1;PDBTitle: the crystal structure of tumor endothelial marker 8 (tem8)2 extracellular domain
19 d1u0oc1
99.2
15
Fold: vWA-likeSuperfamily: vWA-likeFamily: Integrin A (or I) domain20 d1ijba_
99.2
13
Fold: vWA-likeSuperfamily: vWA-likeFamily: Integrin A (or I) domain21 d1atza_
not modelled
99.1
16
Fold: vWA-likeSuperfamily: vWA-likeFamily: Integrin A (or I) domain22 c3gxbB_
not modelled
99.1
13
PDB header: cell adhesionChain: B: PDB Molecule: von willebrand factor;PDBTitle: crystal structure of vwf a2 domain
23 c1jeyB_
not modelled
99.1
17
PDB header: dna binding protein/dnaChain: B: PDB Molecule: ku80;PDBTitle: crystal structure of the ku heterodimer bound to dna
24 d1v7pc_
not modelled
99.1
18
Fold: vWA-likeSuperfamily: vWA-likeFamily: Integrin A (or I) domain25 d1n3ya_
not modelled
99.1
14
Fold: vWA-likeSuperfamily: vWA-likeFamily: Integrin A (or I) domain26 d1ck4a_
not modelled
99.1
17
Fold: vWA-likeSuperfamily: vWA-likeFamily: Integrin A (or I) domain27 d1q0pa_
not modelled
99.1
21
Fold: vWA-likeSuperfamily: vWA-likeFamily: Integrin A (or I) domain28 d1pt6a_
not modelled
98.9
16
Fold: vWA-likeSuperfamily: vWA-likeFamily: Integrin A (or I) domain29 d1mf7a_
not modelled
98.9
13
Fold: vWA-likeSuperfamily: vWA-likeFamily: Integrin A (or I) domain30 d1tyeb2
not modelled
98.9
21
Fold: vWA-likeSuperfamily: vWA-likeFamily: Integrin A (or I) domain31 d1mjna_
not modelled
98.6
19
Fold: vWA-likeSuperfamily: vWA-likeFamily: Integrin A (or I) domain32 c3fcuB_
not modelled
98.6
21
PDB header: cell adhesion/blood clottingChain: B: PDB Molecule: integrin beta-3;PDBTitle: structure of headpiece of integrin aiibb3 in open conformation
33 c3ragA_
not modelled
98.5
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of uncharacterized protein aaci_0196 from2 alicyclobacillus acidocaldarius subsp. acidocaldarius dsm 446
34 c1u8cB_
not modelled
98.4
22
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-3;PDBTitle: a novel adaptation of the integrin psi domain revealed from its2 crystal structure
35 c1jeqA_
not modelled
98.2
18
PDB header: dna binding proteinChain: A: PDB Molecule: ku70;PDBTitle: crystal structure of the ku heterodimer
36 c1pd0A_
not modelled
98.0
23
PDB header: transport proteinChain: A: PDB Molecule: protein transport protein sec24;PDBTitle: crystal structure of the copii coat subunit, sec24,2 complexed with a peptide from the snare protein sed53 (yeast syntaxin-5)
37 c3k6sB_
not modelled
97.8
19
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-2;PDBTitle: structure of integrin alphaxbeta2 ectodomain
38 c1m2vB_
not modelled
97.6
26
PDB header: protein transportChain: B: PDB Molecule: protein transport protein sec24;PDBTitle: crystal structure of the yeast sec23/24 heterodimer
39 c3eh2B_
not modelled
97.6
17
PDB header: protein transportChain: B: PDB Molecule: protein transport protein sec24c;PDBTitle: crystal structure of the human copii-coat protein sec24c
40 c3egxB_
not modelled
97.5
18
PDB header: protein transportChain: B: PDB Molecule: protein transport protein sec24a;PDBTitle: crystal structure of the mammalian copii-coat protein2 sec23a/24a complexed with the snare protein sec22b and3 bound to the transport signal sequence of the snare protein4 bet1
41 c3eg9A_
not modelled
97.5
19
PDB header: protein transportChain: A: PDB Molecule: protein transport protein sec23a;PDBTitle: crystal structure of the mammalian copii-coat protein2 sec23/24 bound to the transport signal sequence of membrin
42 d2qtva3
not modelled
97.4
19
Fold: vWA-likeSuperfamily: vWA-likeFamily: Trunk domain of Sec23/2443 c3eg9B_
not modelled
97.3
16
PDB header: protein transportChain: B: PDB Molecule: sec24 related gene family, member d;PDBTitle: crystal structure of the mammalian copii-coat protein2 sec23/24 bound to the transport signal sequence of membrin
44 c1m2oA_
not modelled
97.0
18
PDB header: protein transport/signaling proteinChain: A: PDB Molecule: protein transport protein sec23;PDBTitle: crystal structure of the sec23-sar1 complex
45 c3eyaE_
not modelled
84.0
13
PDB header: oxidoreductaseChain: E: PDB Molecule: pyruvate dehydrogenase [cytochrome];PDBTitle: structural basis for membrane binding and catalytic2 activation of the peripheral membrane enzyme pyruvate3 oxidase from escherichia coli
46 d2djia3
not modelled
77.6
14
Fold: Thiamin diphosphate-binding fold (THDP-binding)Superfamily: Thiamin diphosphate-binding fold (THDP-binding)Family: Pyruvate oxidase and decarboxylase PP module47 d1jsca3
not modelled
73.7
18
Fold: Thiamin diphosphate-binding fold (THDP-binding)Superfamily: Thiamin diphosphate-binding fold (THDP-binding)Family: Pyruvate oxidase and decarboxylase PP module48 c1jscA_
not modelled
67.6
18
PDB header: lyaseChain: A: PDB Molecule: acetohydroxy-acid synthase;PDBTitle: crystal structure of the catalytic subunit of yeast2 acetohydroxyacid synthase: a target for herbicidal3 inhibitors
49 d1twda_
not modelled
64.0
19
Fold: TIM beta/alpha-barrelSuperfamily: CutC-likeFamily: CutC-like50 d2ji7a3
not modelled
63.6
18
Fold: Thiamin diphosphate-binding fold (THDP-binding)Superfamily: Thiamin diphosphate-binding fold (THDP-binding)Family: Pyruvate oxidase and decarboxylase PP module51 d2piaa2
not modelled
61.7
13
Fold: Ferredoxin reductase-like, C-terminal NADP-linked domainSuperfamily: Ferredoxin reductase-like, C-terminal NADP-linked domainFamily: Aromatic dioxygenase reductase-like52 d1li4a2
not modelled
56.9
21
Fold: Flavodoxin-likeSuperfamily: Formate/glycerate dehydrogenase catalytic domain-likeFamily: S-adenosylhomocystein hydrolase53 d1t9ba3
not modelled
54.3
18
Fold: Thiamin diphosphate-binding fold (THDP-binding)Superfamily: Thiamin diphosphate-binding fold (THDP-binding)Family: Pyruvate oxidase and decarboxylase PP module54 d1dkua2
not modelled
54.1
18
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosylpyrophosphate synthetase-like55 d1ozha3
not modelled
51.7
17
Fold: Thiamin diphosphate-binding fold (THDP-binding)Superfamily: Thiamin diphosphate-binding fold (THDP-binding)Family: Pyruvate oxidase and decarboxylase PP module56 c3iwpK_
not modelled
51.1
16
PDB header: metal binding proteinChain: K: PDB Molecule: copper homeostasis protein cutc homolog;PDBTitle: crystal structure of human copper homeostasis protein cutc
57 d1ybha3
not modelled
49.9
16
Fold: Thiamin diphosphate-binding fold (THDP-binding)Superfamily: Thiamin diphosphate-binding fold (THDP-binding)Family: Pyruvate oxidase and decarboxylase PP module58 d1wd5a_
not modelled
46.4
17
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)59 c2bdqA_
not modelled
46.2
17
PDB header: metal transportChain: A: PDB Molecule: copper homeostasis protein cutc;PDBTitle: crystal structure of the putative copper homeostasis2 protein cutc from streptococcus agalactiae, northeast3 strucural genomics target sar15.
60 d1y0ba1
not modelled
44.7
16
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)61 c3d64A_
not modelled
44.3
19
PDB header: hydrolaseChain: A: PDB Molecule: adenosylhomocysteinase;PDBTitle: crystal structure of s-adenosyl-l-homocysteine hydrolase from2 burkholderia pseudomallei
62 c3oneA_
not modelled
42.3
17
PDB header: hydrolase/hydrolase substrateChain: A: PDB Molecule: adenosylhomocysteinase;PDBTitle: crystal structure of lupinus luteus s-adenosyl-l-homocysteine2 hydrolase in complex with adenine
63 c2yzkC_
not modelled
40.2
17
PDB header: transferaseChain: C: PDB Molecule: orotate phosphoribosyltransferase;PDBTitle: crystal structure of orotate phosphoribosyltransferase from2 aeropyrum pernix
64 d2ez9a3
not modelled
39.6
13
Fold: Thiamin diphosphate-binding fold (THDP-binding)Superfamily: Thiamin diphosphate-binding fold (THDP-binding)Family: Pyruvate oxidase and decarboxylase PP module65 c2vf7B_
not modelled
38.6
20
PDB header: dna-binding proteinChain: B: PDB Molecule: excinuclease abc, subunit a.;PDBTitle: crystal structure of uvra2 from deinococcus radiodurans
66 c2wnsB_
not modelled
37.1
19
PDB header: transferaseChain: B: PDB Molecule: orotate phosphoribosyltransferase;PDBTitle: human orotate phosphoribosyltransferase (oprtase) domain of2 uridine 5'-monophosphate synthase (umps) in complex with3 its substrate orotidine 5'-monophosphate (omp)
67 d2aeea1
not modelled
37.1
16
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)68 c2djiA_
not modelled
36.9
14
PDB header: oxidoreductaseChain: A: PDB Molecule: pyruvate oxidase;PDBTitle: crystal structure of pyruvate oxidase from aerococcus2 viridans containing fad
69 c1d4fD_
not modelled
36.7
17
PDB header: hydrolaseChain: D: PDB Molecule: s-adenosylhomocysteine hydrolase;PDBTitle: crystal structure of recombinant rat-liver d244e mutant s-2 adenosylhomocysteine hydrolase
70 c3m3hA_
not modelled
34.9
19
PDB header: transferaseChain: A: PDB Molecule: orotate phosphoribosyltransferase;PDBTitle: 1.75 angstrom resolution crystal structure of an orotate2 phosphoribosyltransferase from bacillus anthracis str. 'ames3 ancestor'
71 d2r8oa2
not modelled
34.2
14
Fold: Thiamin diphosphate-binding fold (THDP-binding)Superfamily: Thiamin diphosphate-binding fold (THDP-binding)Family: TK-like PP module72 c3pihA_
not modelled
33.7
16
PDB header: hydrolase/dnaChain: A: PDB Molecule: uvrabc system protein a;PDBTitle: t. maritima uvra in complex with fluorescein-modified dna
73 c2jlaD_
not modelled
32.2
12
PDB header: transferaseChain: D: PDB Molecule: 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexenePDBTitle: crystal structure of e.coli mend, 2-succinyl-5-enolpyruvyl-2 6-hydroxy-3-cyclohexadiene-1-carboxylate synthase - semet3 protein
74 d1l1qa_
not modelled
31.3
13
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)75 c2pgnA_
not modelled
31.3
13
PDB header: hydrolaseChain: A: PDB Molecule: cyclohexane-1,2-dione hydrolase (cdh);PDBTitle: the crystal structure of fad and thdp-dependent cyclohexane-1,2-dione2 hydrolase in complex with cyclohexane-1,2-dione
76 c2ji6B_
not modelled
30.4
20
PDB header: lyaseChain: B: PDB Molecule: oxalyl-coa decarboxylase;PDBTitle: x-ray structure of oxalyl-coa decarboxylase in complex with2 3-deaza-thdp and oxalyl-coa
77 c1sy7B_
not modelled
29.4
14
PDB header: oxidoreductaseChain: B: PDB Molecule: catalase 1;PDBTitle: crystal structure of the catalase-1 from neurospora crassa, native2 structure at 1.75a resolution.
78 c1upaC_
not modelled
29.2
13
PDB header: synthaseChain: C: PDB Molecule: carboxyethylarginine synthase;PDBTitle: carboxyethylarginine synthase from streptomyces2 clavuligerus (semet structure)
79 c3n58D_
not modelled
28.9
19
PDB header: hydrolaseChain: D: PDB Molecule: adenosylhomocysteinase;PDBTitle: crystal structure of s-adenosyl-l-homocysteine hydrolase from brucella2 melitensis in ternary complex with nad and adenosine, orthorhombic3 form
80 c3gvpB_
not modelled
28.6
17
PDB header: hydrolaseChain: B: PDB Molecule: adenosylhomocysteinase 3;PDBTitle: human sahh-like domain of human adenosylhomocysteinase 3
81 c1ozhD_
not modelled
28.4
17
PDB header: lyaseChain: D: PDB Molecule: acetolactate synthase, catabolic;PDBTitle: the crystal structure of klebsiella pneumoniae acetolactate2 synthase with enzyme-bound cofactor and with an unusual3 intermediate.
82 d1pvda3
not modelled
26.7
13
Fold: Thiamin diphosphate-binding fold (THDP-binding)Superfamily: Thiamin diphosphate-binding fold (THDP-binding)Family: Pyruvate oxidase and decarboxylase PP module83 c1j9zB_
not modelled
26.0
12
PDB header: oxidoreductaseChain: B: PDB Molecule: nadph-cytochrome p450 reductase;PDBTitle: cypor-w677g
84 c1t9dB_
not modelled
25.9
18
PDB header: transferaseChain: B: PDB Molecule: acetolactate synthase, mitochondrial;PDBTitle: crystal structure of yeast acetohydroxyacid synthase in2 complex with a sulfonylurea herbicide, metsulfuron methyl
85 c2hgsA_
not modelled
25.4
13
PDB header: amine/carboxylate ligaseChain: A: PDB Molecule: protein (glutathione synthetase);PDBTitle: human glutathione synthetase
86 d1q6za3
not modelled
25.2
20
Fold: Thiamin diphosphate-binding fold (THDP-binding)Superfamily: Thiamin diphosphate-binding fold (THDP-binding)Family: Pyruvate oxidase and decarboxylase PP module87 c3gbcA_
not modelled
24.6
10
PDB header: hydrolaseChain: A: PDB Molecule: pyrazinamidase/nicotinamidas pnca;PDBTitle: determination of the crystal structure of the pyrazinamidase from2 m.tuberculosis : a structure-function analysis for prediction3 resistance to pyrazinamide
88 c1yi1A_
not modelled
24.5
14
PDB header: transferaseChain: A: PDB Molecule: acetolactate synthase;PDBTitle: crystal structure of arabidopsis thaliana acetohydroxyacid synthase in2 complex with a sulfonylurea herbicide, tribenuron methyl
89 d1chda_
not modelled
24.4
8
Fold: Methylesterase CheB, C-terminal domainSuperfamily: Methylesterase CheB, C-terminal domainFamily: Methylesterase CheB, C-terminal domain90 d1v8ba2
not modelled
23.6
15
Fold: Flavodoxin-likeSuperfamily: Formate/glycerate dehydrogenase catalytic domain-likeFamily: S-adenosylhomocystein hydrolase91 c3efhB_
not modelled
23.2
19
PDB header: transferaseChain: B: PDB Molecule: ribose-phosphate pyrophosphokinase 1;PDBTitle: crystal structure of human phosphoribosyl pyrophosphate2 synthetase 1
92 d1itza1
not modelled
22.7
11
Fold: Thiamin diphosphate-binding fold (THDP-binding)Superfamily: Thiamin diphosphate-binding fold (THDP-binding)Family: TK-like PP module93 c2panF_
not modelled
22.4
15
PDB header: lyaseChain: F: PDB Molecule: glyoxylate carboligase;PDBTitle: crystal structure of e. coli glyoxylate carboligase
94 c4a2iV_
not modelled
22.2
12
PDB header: ribosome/hydrolaseChain: V: PDB Molecule: putative ribosome biogenesis gtpase rsga;PDBTitle: cryo-electron microscopy structure of the 30s subunit in complex with2 the yjeq biogenesis factor
95 c2bpoA_
not modelled
21.7
13
PDB header: reductaseChain: A: PDB Molecule: nadph-cytochrom p450 reductase;PDBTitle: crystal structure of the yeast cpr triple mutant: d74g,2 y75f, k78a.
96 c2c4kD_
not modelled
21.4
11
PDB header: regulatory proteinChain: D: PDB Molecule: phosphoribosyl pyrophosphate synthetase-PDBTitle: crystal structure of human phosphoribosylpyrophosphate2 synthetase-associated protein 39 (pap39)
97 c1cqxB_
not modelled
21.3
11
PDB header: lipid binding proteinChain: B: PDB Molecule: flavohemoprotein;PDBTitle: crystal structure of the flavohemoglobin from alcaligenes eutrophus at2 1.75 a resolution
98 c3sftA_
not modelled
21.2
11
PDB header: hydrolaseChain: A: PDB Molecule: chemotaxis response regulator protein-glutamatePDBTitle: crystal structure of thermotoga maritima cheb methylesterase catalytic2 domain
99 c2x7jA_
not modelled
21.2
16
PDB header: transferaseChain: A: PDB Molecule: 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexenePDBTitle: structure of the menaquinone biosynthesis protein mend from2 bacillus subtilis
100 c3cueM_
not modelled
21.2
6
PDB header: protein transportChain: M: PDB Molecule: transport protein particle 23 kda subunit;PDBTitle: crystal structure of a trapp subassembly activating the rab2 ypt1p
101 c2dy0A_
not modelled
20.8
20
PDB header: transferaseChain: A: PDB Molecule: adenine phosphoribosyltransferase;PDBTitle: crystal structure of project jw0458 from escherichia coli
102 d1gpua1
not modelled
20.3
15
Fold: Thiamin diphosphate-binding fold (THDP-binding)Superfamily: Thiamin diphosphate-binding fold (THDP-binding)Family: TK-like PP module103 c1tllA_
not modelled
20.1
14
PDB header: oxidoreductaseChain: A: PDB Molecule: nitric-oxide synthase, brain;PDBTitle: crystal structure of rat neuronal nitric-oxide synthase2 reductase module at 2.3 a resolution.