1 c2hwgA_
100.0
97
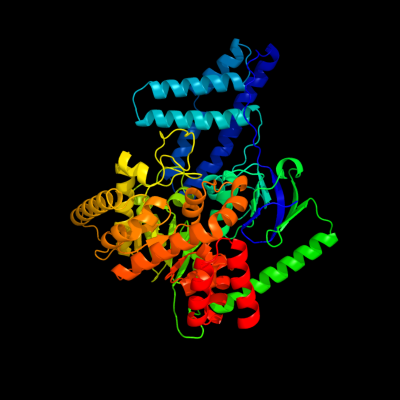
PDB header: transferaseChain: A: PDB Molecule: phosphoenolpyruvate-protein phosphotransferase;PDBTitle: structure of phosphorylated enzyme i of the2 phosphoenolpyruvate:sugar phosphotransferase system
2 c2hroA_
100.0
52
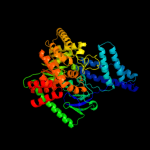
PDB header: transferaseChain: A: PDB Molecule: phosphoenolpyruvate-protein phosphotransferase;PDBTitle: structure of the full-lenght enzyme i of the pts system from2 staphylococcus carnosus
3 c1h6zA_
100.0
23
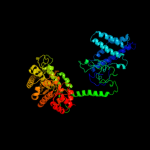
PDB header: transferaseChain: A: PDB Molecule: pyruvate phosphate dikinase;PDBTitle: 3.0 a resolution crystal structure of glycosomal pyruvate2 phosphate dikinase from trypanosoma brucei
4 c1kblA_
100.0
26
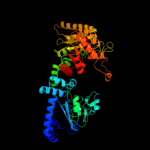
PDB header: transferaseChain: A: PDB Molecule: pyruvate phosphate dikinase;PDBTitle: pyruvate phosphate dikinase
5 c2bg5C_
100.0
58
PDB header: transferaseChain: C: PDB Molecule: phosphoenolpyruvate-protein kinase;PDBTitle: crystal structure of the phosphoenolpyruvate-binding enzyme2 i-domain from the thermoanaerobacter tengcongensis pep:3 sugar phosphotransferase system (pts)
6 c1vbhA_
100.0
24
PDB header: transferaseChain: A: PDB Molecule: pyruvate,orthophosphate dikinase;PDBTitle: pyruvate phosphate dikinase with bound mg-pep from maize
7 c2olsA_
100.0
27
PDB header: transferaseChain: A: PDB Molecule: phosphoenolpyruvate synthase;PDBTitle: the crystal structure of the phosphoenolpyruvate synthase from2 neisseria meningitidis
8 d1h6za1
100.0
28
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Pyruvate phosphate dikinase, C-terminal domain9 d1vbga1
100.0
29
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Pyruvate phosphate dikinase, C-terminal domain10 d1kbla1
100.0
31
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Pyruvate phosphate dikinase, C-terminal domain11 c1ezaA_
100.0
100
PDB header: phosphotransferaseChain: A: PDB Molecule: enzyme i;PDBTitle: amino terminal domain of enzyme i from escherichia coli nmr,2 restrained regularized mean structure
12 d1zyma2
99.9
96
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: Phosphohistidine domainFamily: N-terminal domain of enzyme I of the PEP:sugar phosphotransferase system13 d1zyma1
99.9
100
Fold: SAM domain-likeSuperfamily: Enzyme I of the PEP:sugar phosphotransferase system HPr-binding (sub)domainFamily: Enzyme I of the PEP:sugar phosphotransferase system HPr-binding (sub)domain14 d1dxea_
99.9
19
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: HpcH/HpaI aldolase15 c3qz6A_
99.9
22
PDB header: lyaseChain: A: PDB Molecule: hpch/hpai aldolase;PDBTitle: the crystal structure of hpch/hpai aldolase from desulfitobacterium2 hafniense dcb-2
16 d1izca_
99.8
21
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: HpcH/HpaI aldolase17 c1izcA_
99.8
21
PDB header: lyaseChain: A: PDB Molecule: macrophomate synthase intermolecular diels-alderase;PDBTitle: crystal structure analysis of macrophomate synthase
18 d1vbga2
99.8
21
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: Phosphohistidine domainFamily: Pyruvate phosphate dikinase, central domain19 c2v5jB_
99.8
24
PDB header: lyaseChain: B: PDB Molecule: 2,4-dihydroxyhept-2-ene-1,7-dioic acid aldolase;PDBTitle: apo class ii aldolase hpch
20 d1h6za2
99.8
23
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: Phosphohistidine domainFamily: Pyruvate phosphate dikinase, central domain21 d1kbla2
not modelled
99.8
26
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: Phosphohistidine domainFamily: Pyruvate phosphate dikinase, central domain22 c2vwtA_
not modelled
99.8
21
PDB header: lyaseChain: A: PDB Molecule: yfau, 2-keto-3-deoxy sugar aldolase;PDBTitle: crystal structure of yfau, a metal ion dependent class ii2 aldolase from escherichia coli k12 - mg-pyruvate product3 complex
23 c3t07D_
not modelled
99.5
18
PDB header: transferase/transferase inhibitorChain: D: PDB Molecule: pyruvate kinase;PDBTitle: crystal structure of s. aureus pyruvate kinase in complex with a2 naturally occurring bis-indole alkaloid
24 c2e28A_
not modelled
99.4
28
PDB header: transferaseChain: A: PDB Molecule: pyruvate kinase;PDBTitle: crystal structure analysis of pyruvate kinase from bacillus2 stearothermophilus
25 c3qqwC_
not modelled
99.4
15
PDB header: lyaseChain: C: PDB Molecule: putative citrate lyase;PDBTitle: crystal structure of a hypothetical lyase (reut_b4148) from ralstonia2 eutropha jmp134 at 2.44 a resolution
26 c1sgjB_
not modelled
99.3
21
PDB header: lyaseChain: B: PDB Molecule: citrate lyase, beta subunit;PDBTitle: crystal structure of citrate lyase beta subunit
27 d1sgja_
not modelled
99.3
21
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: HpcH/HpaI aldolase28 d1e0ta2
not modelled
99.2
14
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Pyruvate kinase29 d2g50a2
not modelled
99.2
16
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Pyruvate kinase30 d1a3xa2
not modelled
99.2
17
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Pyruvate kinase31 d1pkla2
not modelled
99.1
19
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Pyruvate kinase32 c3r4iB_
not modelled
99.0
27
PDB header: lyaseChain: B: PDB Molecule: citrate lyase;PDBTitle: crystal structure of a citrate lyase (bxe_b2899) from burkholderia2 xenovorans lb400 at 2.24 a resolution
33 d1liua2
not modelled
99.0
16
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Pyruvate kinase34 c1u5vA_
not modelled
99.0
18
PDB header: lyaseChain: A: PDB Molecule: cite;PDBTitle: structure of cite complexed with triphosphate group of atp2 form mycobacterium tuberculosis
35 d1u5ha_
not modelled
99.0
19
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: HpcH/HpaI aldolase36 c3cuzA_
not modelled
98.8
13
PDB header: transferaseChain: A: PDB Molecule: malate synthase a;PDBTitle: atomic resolution structures of escherichia coli and2 bacillis anthracis malate synthase a: comparison with3 isoform g and implications for structure based drug design
37 c3qllB_
not modelled
98.7
14
PDB header: lyaseChain: B: PDB Molecule: citrate lyase;PDBTitle: crystal structure of ripc from yersinia pestis
38 c3cuxA_
not modelled
98.3
16
PDB header: transferaseChain: A: PDB Molecule: malate synthase;PDBTitle: atomic resolution structures of escherichia coli and2 bacillis anthracis malate synthase a: comparison with3 isoform g and implications for structure based drug design
39 c2vgbB_
not modelled
98.3
18
PDB header: transferaseChain: B: PDB Molecule: pyruvate kinase isozymes r/l;PDBTitle: human erythrocyte pyruvate kinase
40 c3ma8A_
not modelled
98.1
17
PDB header: transferaseChain: A: PDB Molecule: pyruvate kinase;PDBTitle: crystal structure of cgd1_2040, a pyruvate kinase from cryptosporidium2 parvum
41 c3eoeC_
not modelled
98.1
16
PDB header: transferaseChain: C: PDB Molecule: pyruvate kinase;PDBTitle: crystal structure of pyruvate kinase from toxoplasma gondii, 55.m00007
42 c3pugA_
not modelled
97.9
17
PDB header: transferaseChain: A: PDB Molecule: malate synthase;PDBTitle: haloferax volcanii malate synthase native at 3mm glyoxylate
43 c1e0tD_
not modelled
97.9
15
PDB header: phosphotransferaseChain: D: PDB Molecule: pyruvate kinase;PDBTitle: r292d mutant of e. coli pyruvate kinase
44 c1aqfB_
not modelled
97.7
15
PDB header: transferaseChain: B: PDB Molecule: pyruvate kinase;PDBTitle: pyruvate kinase from rabbit muscle with mg, k, and l-2 phospholactate
45 c1t5aB_
not modelled
97.7
17
PDB header: transferaseChain: B: PDB Molecule: pyruvate kinase, m2 isozyme;PDBTitle: human pyruvate kinase m2
46 c3e0vB_
not modelled
97.6
20
PDB header: transferaseChain: B: PDB Molecule: pyruvate kinase;PDBTitle: crystal structure of pyruvate kinase from leishmania mexicana in2 complex with sulphate ions
47 c1a3wB_
not modelled
97.6
18
PDB header: transferaseChain: B: PDB Molecule: pyruvate kinase;PDBTitle: pyruvate kinase from saccharomyces cerevisiae complexed with fbp, pg,2 mn2+ and k+
48 c1pklB_
not modelled
97.6
20
PDB header: transferaseChain: B: PDB Molecule: protein (pyruvate kinase);PDBTitle: the structure of leishmania pyruvate kinase
49 d1d8ca_
not modelled
97.5
14
Fold: TIM beta/alpha-barrelSuperfamily: Malate synthase GFamily: Malate synthase G50 c3khdC_
not modelled
96.7
24
PDB header: transferaseChain: C: PDB Molecule: pyruvate kinase;PDBTitle: crystal structure of pff1300w.
51 c2zbtB_
not modelled
95.3
21
PDB header: lyaseChain: B: PDB Molecule: pyridoxal biosynthesis lyase pdxs;PDBTitle: crystal structure of pyridoxine biosynthesis protein from thermus2 thermophilus hb8
52 c2nv2U_
not modelled
94.8
23
PDB header: lyase/transferaseChain: U: PDB Molecule: pyridoxal biosynthesis lyase pdxs;PDBTitle: structure of the plp synthase complex pdx1/2 (yaad/e) from bacillus2 subtilis
53 c3femB_
not modelled
94.6
22
PDB header: biosynthetic protein, transferaseChain: B: PDB Molecule: pyridoxine biosynthesis protein snz1;PDBTitle: structure of the synthase subunit pdx1.1 (snz1) of plp synthase from2 saccharomyces cerevisiae
54 d1eepa_
not modelled
94.4
20
Fold: TIM beta/alpha-barrelSuperfamily: Inosine monophosphate dehydrogenase (IMPDH)Family: Inosine monophosphate dehydrogenase (IMPDH)55 c2a7rD_
not modelled
93.6
18
PDB header: oxidoreductaseChain: D: PDB Molecule: gmp reductase 2;PDBTitle: crystal structure of human guanosine monophosphate2 reductase 2 (gmpr2)
56 d1vrda1
not modelled
92.7
21
Fold: TIM beta/alpha-barrelSuperfamily: Inosine monophosphate dehydrogenase (IMPDH)Family: Inosine monophosphate dehydrogenase (IMPDH)57 d1jr1a1
not modelled
92.0
16
Fold: TIM beta/alpha-barrelSuperfamily: Inosine monophosphate dehydrogenase (IMPDH)Family: Inosine monophosphate dehydrogenase (IMPDH)58 c3odmE_
not modelled
92.0
15
PDB header: lyaseChain: E: PDB Molecule: phosphoenolpyruvate carboxylase;PDBTitle: archaeal-type phosphoenolpyruvate carboxylase
59 d1n8ia_
not modelled
91.9
13
Fold: TIM beta/alpha-barrelSuperfamily: Malate synthase GFamily: Malate synthase G60 d2hi6a1
not modelled
91.7
25
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: LeuD/IlvD-likeFamily: AF0055-like61 c3r2gA_
not modelled
91.0
18
PDB header: oxidoreductaseChain: A: PDB Molecule: inosine 5'-monophosphate dehydrogenase;PDBTitle: crystal structure of inosine 5' monophosphate dehydrogenase from2 legionella pneumophila
62 c1vrdA_
not modelled
90.8
21
PDB header: oxidoreductaseChain: A: PDB Molecule: inosine-5'-monophosphate dehydrogenase;PDBTitle: crystal structure of inosine-5'-monophosphate dehydrogenase (tm1347)2 from thermotoga maritima at 2.18 a resolution
63 d1zfja1
not modelled
89.7
16
Fold: TIM beta/alpha-barrelSuperfamily: Inosine monophosphate dehydrogenase (IMPDH)Family: Inosine monophosphate dehydrogenase (IMPDH)64 c3khjE_
not modelled
88.5
22
PDB header: oxidoreductaseChain: E: PDB Molecule: inosine-5-monophosphate dehydrogenase;PDBTitle: c. parvum inosine monophosphate dehydrogenase bound by inhibitor c64
65 c1jcnA_
not modelled
87.7
18
PDB header: oxidoreductaseChain: A: PDB Molecule: inosine monophosphate dehydrogenase i;PDBTitle: binary complex of human type-i inosine monophosphate dehydrogenase2 with 6-cl-imp
66 d1tb3a1
not modelled
87.6
24
Fold: TIM beta/alpha-barrelSuperfamily: FMN-linked oxidoreductasesFamily: FMN-linked oxidoreductases67 c3qjaA_
not modelled
86.9
16
PDB header: lyaseChain: A: PDB Molecule: indole-3-glycerol phosphate synthase;PDBTitle: crystal structure of the mycobacterium tuberculosis indole-3-glycerol2 phosphate synthase (trpc) in apo form
68 d1pvna1
not modelled
86.7
15
Fold: TIM beta/alpha-barrelSuperfamily: Inosine monophosphate dehydrogenase (IMPDH)Family: Inosine monophosphate dehydrogenase (IMPDH)69 c3pm6B_
not modelled
86.1
13
PDB header: lyaseChain: B: PDB Molecule: putative fructose-bisphosphate aldolase;PDBTitle: crystal structure of a putative fructose-1,6-biphosphate aldolase from2 coccidioides immitis solved by combined sad mr
70 d1jcna1
not modelled
83.3
20
Fold: TIM beta/alpha-barrelSuperfamily: Inosine monophosphate dehydrogenase (IMPDH)Family: Inosine monophosphate dehydrogenase (IMPDH)71 c2yzrB_
not modelled
82.9
21
PDB header: lyaseChain: B: PDB Molecule: pyridoxal biosynthesis lyase pdxs;PDBTitle: crystal structure of pyridoxine biosynthesis protein from2 methanocaldococcus jannaschii
72 c2vc6A_
not modelled
82.3
26
PDB header: lyaseChain: A: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: structure of mosa from s. meliloti with pyruvate bound
73 d2cu0a1
not modelled
79.8
22
Fold: TIM beta/alpha-barrelSuperfamily: Inosine monophosphate dehydrogenase (IMPDH)Family: Inosine monophosphate dehydrogenase (IMPDH)74 d2gp4a1
not modelled
77.4
19
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: LeuD/IlvD-likeFamily: IlvD/EDD C-terminal domain-like75 c1ypfB_
not modelled
77.1
18
PDB header: oxidoreductaseChain: B: PDB Molecule: gmp reductase;PDBTitle: crystal structure of guac (ba5705) from bacillus anthracis at 1.8 a2 resolution
76 c2e77B_
not modelled
75.2
17
PDB header: oxidoreductaseChain: B: PDB Molecule: lactate oxidase;PDBTitle: crystal structure of l-lactate oxidase with pyruvate complex
77 c2zrvC_
not modelled
74.9
22
PDB header: isomeraseChain: C: PDB Molecule: isopentenyl-diphosphate delta-isomerase;PDBTitle: crystal structure of sulfolobus shibatae isopentenyl2 diphosphate isomerase in complex with reduced fmn.
78 d1goxa_
not modelled
74.7
24
Fold: TIM beta/alpha-barrelSuperfamily: FMN-linked oxidoreductasesFamily: FMN-linked oxidoreductases79 d1o5ka_
not modelled
74.5
24
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase80 c2f7fA_
not modelled
73.7
17
PDB header: transferaseChain: A: PDB Molecule: nicotinate phosphoribosyltransferase, putative;PDBTitle: crystal structure of enterococcus faecalis putative nicotinate2 phosphoribosyltransferase, new york structural genomics consortium
81 c2rduA_
not modelled
72.3
18
PDB header: oxidoreductaseChain: A: PDB Molecule: hydroxyacid oxidase 1;PDBTitle: crystal structure of human glycolate oxidase in complex with2 glyoxylate
82 c2gp4A_
not modelled
71.8
21
PDB header: lyaseChain: A: PDB Molecule: 6-phosphogluconate dehydratase;PDBTitle: structure of [fes]cluster-free apo form of 6-phosphogluconate2 dehydratase from shewanella oneidensis
83 c3n2xB_
not modelled
70.9
15
PDB header: lyaseChain: B: PDB Molecule: uncharacterized protein yage;PDBTitle: crystal structure of yage, a prophage protein belonging to the2 dihydrodipicolinic acid synthase family from e. coli k12 in complex3 with pyruvate
84 d1j93a_
not modelled
70.8
12
Fold: TIM beta/alpha-barrelSuperfamily: UROD/MetE-likeFamily: Uroporphyrinogen decarboxylase, UROD85 c2qr6A_
not modelled
69.4
20
PDB header: oxidoreductaseChain: A: PDB Molecule: imp dehydrogenase/gmp reductase;PDBTitle: crystal structure of imp dehydrogenase/gmp reductase-like protein2 (np_599840.1) from corynebacterium glutamicum atcc 13032 kitasato at3 1.50 a resolution
86 c1me9A_
not modelled
68.7
16
PDB header: oxidoreductaseChain: A: PDB Molecule: inosine-5'-monophosphate dehydrogenase;PDBTitle: inosine monophosphate dehydrogenase (impdh) from2 tritrichomonas foetus with imp bound
87 c3s5oA_
not modelled
68.6
14
PDB header: lyaseChain: A: PDB Molecule: 4-hydroxy-2-oxoglutarate aldolase, mitochondrial;PDBTitle: crystal structure of human 4-hydroxy-2-oxoglutarate aldolase bound to2 pyruvate
88 c3lerA_
not modelled
68.5
23
PDB header: lyaseChain: A: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from2 campylobacter jejuni subsp. jejuni nctc 11168
89 d1a53a_
not modelled
68.3
16
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes90 c3pueA_
not modelled
68.3
21
PDB header: lyaseChain: A: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of the complex of dhydrodipicolinate synthase from2 acinetobacter baumannii with lysine at 2.6a resolution
91 d1qapa1
not modelled
67.9
14
Fold: TIM beta/alpha-barrelSuperfamily: Nicotinate/Quinolinate PRTase C-terminal domain-likeFamily: NadC C-terminal domain-like92 c2cu0B_
not modelled
66.8
22
PDB header: oxidoreductaseChain: B: PDB Molecule: inosine-5'-monophosphate dehydrogenase;PDBTitle: crystal structure of inosine-5'-monophosphate dehydrogenase from2 pyrococcus horikoshii ot3
93 c3lciA_
not modelled
64.8
20
PDB header: lyaseChain: A: PDB Molecule: n-acetylneuraminate lyase;PDBTitle: the d-sialic acid aldolase mutant v251w
94 c2ehhE_
not modelled
63.8
15
PDB header: lyaseChain: E: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from2 aquifex aeolicus
95 d1hl2a_
not modelled
62.9
18
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase96 c3na8A_
not modelled
62.5
24
PDB header: lyaseChain: A: PDB Molecule: putative dihydrodipicolinate synthetase;PDBTitle: crystal structure of a putative dihydrodipicolinate synthetase from2 pseudomonas aeruginosa
97 c2a7nA_
not modelled
61.5
19
PDB header: oxidoreductaseChain: A: PDB Molecule: l(+)-mandelate dehydrogenase;PDBTitle: crystal structure of the g81a mutant of the active chimera of (s)-2 mandelate dehydrogenase
98 c3dz1A_
not modelled
60.4
14
PDB header: lyaseChain: A: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 1.87a resolution
99 c2c3zA_
not modelled
60.1
18
PDB header: lyaseChain: A: PDB Molecule: indole-3-glycerol phosphate synthase;PDBTitle: crystal structure of a truncated variant of indole-3-2 glycerol phosphate synthase from sulfolobus solfataricus
100 c2pkpA_
not modelled
60.0
17
PDB header: lyaseChain: A: PDB Molecule: homoaconitase small subunit;PDBTitle: crystal structure of 3-isopropylmalate dehydratase (leud)2 from methhanocaldococcus jannaschii dsm2661 (mj1271)
101 c3cprB_
not modelled
58.7
25
PDB header: lyaseChain: B: PDB Molecule: dihydrodipicolinate synthetase;PDBTitle: the crystal structure of corynebacterium glutamicum2 dihydrodipicolinate synthase to 2.2 a resolution
102 d1xxxa1
not modelled
58.1
27
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase103 c3iz5H_
not modelled
57.9
34
PDB header: ribosomeChain: H: PDB Molecule: 60s ribosomal protein l7a (l7ae);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
104 d1qpoa1
not modelled
57.6
17
Fold: TIM beta/alpha-barrelSuperfamily: Nicotinate/Quinolinate PRTase C-terminal domain-likeFamily: NadC C-terminal domain-like105 c2rfgB_
not modelled
57.2
21
PDB header: lyaseChain: B: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from hahella2 chejuensis at 1.5a resolution
106 c2r8wB_
not modelled
56.9
18
PDB header: lyaseChain: B: PDB Molecule: agr_c_1641p;PDBTitle: the crystal structure of dihydrodipicolinate synthase (atu0899) from2 agrobacterium tumefaciens str. c58
107 c2v9dB_
not modelled
56.4
15
PDB header: lyaseChain: B: PDB Molecule: yage;PDBTitle: crystal structure of yage, a prophage protein belonging to2 the dihydrodipicolinic acid synthase family from e. coli3 k12
108 c3daqB_
not modelled
55.6
15
PDB header: lyaseChain: B: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from methicillin-2 resistant staphylococcus aureus
109 c1zfjA_
not modelled
55.0
16
PDB header: oxidoreductaseChain: A: PDB Molecule: inosine monophosphate dehydrogenase;PDBTitle: inosine monophosphate dehydrogenase (impdh; ec 1.1.1.205) from2 streptococcus pyogenes
110 d1v7la_
not modelled
55.0
13
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: LeuD/IlvD-likeFamily: LeuD-like111 c1kbiB_
not modelled
54.6
24
PDB header: oxidoreductaseChain: B: PDB Molecule: cytochrome b2;PDBTitle: crystallographic study of the recombinant flavin-binding domain of2 baker's yeast flavocytochrome b2: comparison with the intact wild-3 type enzyme
112 c3ke8A_
not modelled
54.2
17
PDB header: oxidoreductaseChain: A: PDB Molecule: 4-hydroxy-3-methylbut-2-enyl diphosphatePDBTitle: crystal structure of isph:hmbpp-complex
113 c2yxgD_
not modelled
54.1
18
PDB header: lyaseChain: D: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihyrodipicolinate synthase (dapa)
114 c2cdh1_
not modelled
54.0
18
PDB header: transferaseChain: 1: PDB Molecule: enoyl reductase;PDBTitle: architecture of the thermomyces lanuginosus fungal fatty2 acid synthase at 5 angstrom resolution.
115 c2zkrf_
not modelled
54.0
29
PDB header: ribosomal protein/rnaChain: F: PDB Molecule: rna expansion segment es7 part iii;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
116 c1x1oC_
not modelled
52.5
17
PDB header: transferaseChain: C: PDB Molecule: nicotinate-nucleotide pyrophosphorylase;PDBTitle: crystal structure of project id tt0268 from thermus thermophilus hb8
117 c2vy9A_
not modelled
52.4
16
PDB header: gene regulationChain: A: PDB Molecule: anti-sigma-factor antagonist;PDBTitle: molecular architecture of the stressosome, a signal2 integration and transduction hub
118 c3noeA_
not modelled
52.1
22
PDB header: lyaseChain: A: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from pseudomonas2 aeruginosa
119 d1o4ua1
not modelled
50.6
25
Fold: TIM beta/alpha-barrelSuperfamily: Nicotinate/Quinolinate PRTase C-terminal domain-likeFamily: NadC C-terminal domain-like120 c3izcH_
not modelled
50.1
29
PDB header: ribosomeChain: H: PDB Molecule: 60s ribosomal protein rpl8 (l7ae);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome