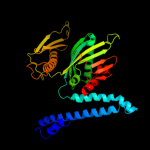
1 d1gqea_
100.0
97
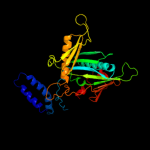
Fold: Release factorSuperfamily: Release factorFamily: Release factor2 c2ihr1_
100.0
46
PDB header: translationChain: 1: PDB Molecule: peptide chain release factor 2;PDBTitle: rf2 of thermus thermophilus
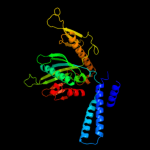
3 c3d5cX_
100.0
34
PDB header: ribosomeChain: X: PDB Molecule: peptide chain release factor 1;PDBTitle: structural basis for translation termination on the 70s ribosome. this2 file contains the 30s subunit, release factor 1 (rf1), two trna, and3 mrna molecules of the second 70s ribosome. the entire crystal4 structure contains two 70s ribosomes as described in remark 400.
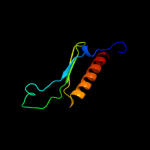
4 d1rq0a_
100.0
39
Fold: Release factorSuperfamily: Release factorFamily: Release factor5 d2b3tb1
100.0
43
Fold: Release factorSuperfamily: Release factorFamily: Release factor6 c1zbtA_
100.0
30
PDB header: translationChain: A: PDB Molecule: peptide chain release factor 1;PDBTitle: crystal structure of peptide chain release factor 1 (rf-1) (smu.1085)2 from streptococcus mutans at 2.34 a resolution
7 c2jvaA_
99.8
26
PDB header: hydrolaseChain: A: PDB Molecule: peptidyl-trna hydrolase domain protein;PDBTitle: nmr solution structure of peptidyl-trna hydrolase domain protein from2 pseudomonas syringae pv. tomato. northeast structural genomics3 consortium target psr211
8 c2jy9A_
99.8
35
PDB header: hydrolaseChain: A: PDB Molecule: putative trna hydrolase domain;PDBTitle: nmr structure of putative trna hydrolase domain from2 salmonella typhimurium. northeast structural genomics3 consortium target str220
9 d1j26a_
99.8
25
Fold: dsRBD-likeSuperfamily: Peptidyl-tRNA hydrolase domain-likeFamily: Peptidyl-tRNA hydrolase domain10 c3errB_
91.6
11
PDB header: ligaseChain: B: PDB Molecule: fusion protein of microtubule binding domain fromPDBTitle: microtubule binding domain from mouse cytoplasmic dynein as2 a fusion with seryl-trna synthetase
11 c1wpaA_
90.3
17
PDB header: cell adhesionChain: A: PDB Molecule: occludin;PDBTitle: 1.5 angstrom crystal structure of human occludin fragment2 413-522
12 c1xawA_
89.7
17
PDB header: cell adhesionChain: A: PDB Molecule: occludin;PDBTitle: crystal structure of the cytoplasmic distal c-terminal2 domain of occludin
13 d1fpoa2
85.8
11
Fold: Open three-helical up-and-down bundleSuperfamily: HSC20 (HSCB), C-terminal oligomerisation domainFamily: HSC20 (HSCB), C-terminal oligomerisation domain14 c3layF_
60.9
22
PDB header: metal binding proteinChain: F: PDB Molecule: zinc resistance-associated protein;PDBTitle: alpha-helical barrel formed by the decamer of the zinc resistance-2 associated protein (stm4172) from salmonella enterica subsp. enterica3 serovar typhimurium str. lt2
15 c2dq3A_
56.2
20
PDB header: ligaseChain: A: PDB Molecule: seryl-trna synthetase;PDBTitle: crystal structure of aq_298
16 c3uo2A_
49.6
13
PDB header: chaperoneChain: A: PDB Molecule: j-type co-chaperone jac1, mitochondrial;PDBTitle: jac1 co-chaperone from saccharomyces cerevisiae
17 c2dq0A_
46.2
13
PDB header: ligaseChain: A: PDB Molecule: seryl-trna synthetase;PDBTitle: crystal structure of seryl-trna synthetase from pyrococcus2 horikoshii complexed with a seryl-adenylate analog
18 c1deqO_
45.6
11
PDB header: PDB COMPND: 19 d1seta1
45.2
20
Fold: Long alpha-hairpinSuperfamily: tRNA-binding armFamily: Seryl-tRNA synthetase (SerRS)20 c3dtpA_
43.7
14
PDB header: contractile proteinChain: A: PDB Molecule: myosin 2 heavy chain chimera of smooth andPDBTitle: tarantula heavy meromyosin obtained by flexible docking to2 tarantula muscle thick filament cryo-em 3d-map
21 c3ghgK_
not modelled
41.7
13
PDB header: blood clottingChain: K: PDB Molecule: fibrinogen beta chain;PDBTitle: crystal structure of human fibrinogen
22 c2wmoA_
not modelled
39.2
14
PDB header: cell cycleChain: A: PDB Molecule: dedicator of cytokinesis protein 9;PDBTitle: structure of the complex between dock9 and cdc42.
23 c2e1mA_
not modelled
38.0
27
PDB header: oxidoreductaseChain: A: PDB Molecule: l-glutamate oxidase;PDBTitle: crystal structure of l-glutamate oxidase from streptomyces sp. x-119-6
24 c2jyaA_
not modelled
37.0
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein atu1810;PDBTitle: nmr solution structure of protein atu1810 from agrobacterium2 tumefaciens. northeast structural genomics consortium target atr23,3 ontario centre for structural proteomics target atc1776
25 c3lssA_
not modelled
34.4
13
PDB header: ligaseChain: A: PDB Molecule: seryl-trna synthetase;PDBTitle: trypanosoma brucei seryl-trna synthetase in complex with atp
26 c1f8sA_
not modelled
33.4
25
PDB header: oxidoreductaseChain: A: PDB Molecule: l-amino acid oxidase;PDBTitle: crystal structure of l-amino acid oxidase from calloselasma2 rhodostoma, complexed with three molecules of o-aminobenzoate.
27 c3s2xB_
not modelled
32.2
19
PDB header: transferaseChain: B: PDB Molecule: acetyl-coa synthase subunit alpha;PDBTitle: structure of acetyl-coenzyme a synthase alpha subunit c-terminal2 domain
28 c3fhnA_
not modelled
32.1
19
PDB header: transport proteinChain: A: PDB Molecule: protein transport protein tip20;PDBTitle: structure of tip20p
29 c2yrvA_
not modelled
30.3
19
PDB header: transcriptionChain: A: PDB Molecule: at-rich interactive domain-containing protein 4a;PDBTitle: solution structure of the rbb1nt domain of human2 rb(retinoblastoma)-binding protein 1
30 c1s1hI_
not modelled
30.2
15
PDB header: ribosomeChain: I: PDB Molecule: 40s ribosomal protein s16;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1h,4 contains 40s subunit. the 60s ribosomal subunit is in file5 1s1i.
31 c1wleB_
not modelled
29.1
21
PDB header: ligaseChain: B: PDB Molecule: seryl-trna synthetase;PDBTitle: crystal structure of mammalian mitochondrial seryl-trna2 synthetase complexed with seryl-adenylate
32 c3qo8A_
not modelled
29.0
7
PDB header: ligaseChain: A: PDB Molecule: seryl-trna synthetase, cytoplasmic;PDBTitle: crystal structure of seryl-trna synthetase from candida albicans
33 d1cuna2
not modelled
28.8
11
Fold: Spectrin repeat-likeSuperfamily: Spectrin repeatFamily: Spectrin repeat34 c2chpC_
not modelled
28.1
19
PDB header: dna-binding proteinChain: C: PDB Molecule: metalloregulation dna-binding stress protein;PDBTitle: crystal structure of the dodecameric ferritin mrga from2 b.subtilis 168
35 d1tjoa_
not modelled
26.3
18
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin36 c1sryB_
not modelled
25.0
15
PDB header: ligase(synthetase)Chain: B: PDB Molecule: seryl-trna synthetase;PDBTitle: refined crystal structure of the seryl-trna synthetase from2 thermus thermophilus at 2.5 angstroms resolution
37 d2spca_
not modelled
24.7
18
Fold: Spectrin repeat-likeSuperfamily: Spectrin repeatFamily: Spectrin repeat38 d1mqva_
not modelled
24.3
8
Fold: Four-helical up-and-down bundleSuperfamily: CytochromesFamily: Cytochrome c'-like39 c1ei3E_
not modelled
23.6
15
PDB header: PDB COMPND: 40 c2gl2B_
not modelled
23.5
7
PDB header: cell adhesionChain: B: PDB Molecule: adhesion a;PDBTitle: crystal structure of the tetra muntant (t66g,r67g,f68g,2 y69g) of bacterial adhesin fada
41 d1ivsa1
not modelled
23.4
15
Fold: Long alpha-hairpinSuperfamily: tRNA-binding armFamily: Valyl-tRNA synthetase (ValRS) C-terminal domain42 d1nj1a3
not modelled
23.1
17
Fold: Class II aaRS and biotin synthetasesSuperfamily: Class II aaRS and biotin synthetasesFamily: Class II aminoacyl-tRNA synthetase (aaRS)-like, catalytic domain43 d1s05a_
not modelled
22.2
12
Fold: Four-helical up-and-down bundleSuperfamily: CytochromesFamily: Cytochrome c'-like44 c3iq1A_
not modelled
21.1
15
PDB header: metal transportChain: A: PDB Molecule: dps family protein;PDBTitle: crystal structure of dps protein from vibrio cholerae o1, a member of2 a broad superfamily of ferritin-like diiron-carboxylate proteins
45 c3gitA_
not modelled
19.9
19
PDB header: transferaseChain: A: PDB Molecule: carbon monoxide dehydrogenase/acetyl-coa synthase subunitPDBTitle: crystal structure of a truncated acetyl-coa synthase
46 c2c41K_
not modelled
19.8
10
PDB header: iron-binding/oxidation proteinChain: K: PDB Molecule: dps family dna-binding stress response protein;PDBTitle: x-ray structure of dps from thermosynechococcus elongatus
47 c2r6cG_
not modelled
19.7
12
PDB header: replicationChain: G: PDB Molecule: dnag primase, helicase binding domain;PDBTitle: crystal form bh2
48 c2cazF_
not modelled
19.4
60
PDB header: protein transportChain: F: PDB Molecule: protein srn2;PDBTitle: escrt-i core
49 c3ojaB_
not modelled
18.8
16
PDB header: protein bindingChain: B: PDB Molecule: anopheles plasmodium-responsive leucine-rich repeat proteinPDBTitle: crystal structure of lrim1/apl1c complex
50 c1s35A_
not modelled
18.5
13
PDB header: structural proteinChain: A: PDB Molecule: spectrin beta chain, erythrocyte;PDBTitle: crystal structure of repeats 8 and 9 of human erythroid2 spectrin
51 c1ei3C_
not modelled
18.2
9
PDB header: PDB COMPND: 52 c2c5iT_
not modelled
18.2
19
PDB header: protein transportChain: T: PDB Molecule: t-snare affecting a late golgi compartmentPDBTitle: n-terminal domain of tlg1 complexed with n-terminus of2 vps51 in distorted conformation
53 d1gqaa_
not modelled
18.0
18
Fold: Four-helical up-and-down bundleSuperfamily: CytochromesFamily: Cytochrome c'-like54 d1s35a1
not modelled
17.5
15
Fold: Spectrin repeat-likeSuperfamily: Spectrin repeatFamily: Spectrin repeat55 c2vvlD_
not modelled
17.4
27
PDB header: oxidoreductaseChain: D: PDB Molecule: monoamine oxidase n;PDBTitle: the structure of mao-n-d3, a variant of monoamine oxidase2 from aspergillus niger.
56 c2qbvA_
not modelled
16.6
14
PDB header: isomeraseChain: A: PDB Molecule: chorismate mutase;PDBTitle: crystal structure of intracellular chorismate mutase from2 mycobacterium tuberculosis
57 d2gy9i1
not modelled
16.6
38
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Translational machinery components58 c3kwoA_
not modelled
16.4
15
PDB header: oxidoreductaseChain: A: PDB Molecule: putative bacterioferritin;PDBTitle: crystal structure of putative bacterioferritin from2 campylobacter jejuni
59 d2vqei1
not modelled
16.2
28
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Translational machinery components60 d2pc6a2
not modelled
15.9
15
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: IlvH-like61 c2d5kC_
not modelled
15.8
12
PDB header: metal binding proteinChain: C: PDB Molecule: dps family protein;PDBTitle: crystal structure of dps from staphylococcus aureus
62 c3ci9B_
not modelled
15.8
11
PDB header: transcriptionChain: B: PDB Molecule: heat shock factor-binding protein 1;PDBTitle: crystal structure of the human hsbp1
63 d1fafa_
not modelled
15.7
18
Fold: Long alpha-hairpinSuperfamily: Chaperone J-domainFamily: Chaperone J-domain64 d1m0da_
not modelled
15.7
33
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Endonuclease I (Holliday junction resolvase)65 c3h0gA_
not modelled
15.7
17
PDB header: transcriptionChain: A: PDB Molecule: dna-directed rna polymerase ii subunit rpb1;PDBTitle: rna polymerase ii from schizosaccharomyces pombe
66 d1ji4a_
not modelled
15.3
17
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin67 d1cpqa_
not modelled
15.2
18
Fold: Four-helical up-and-down bundleSuperfamily: CytochromesFamily: Cytochrome c'-like68 c3m3wA_
not modelled
15.2
11
PDB header: endocytosisChain: A: PDB Molecule: protein kinase c and casein kinase ii substrate protein 3;PDBTitle: crystal strcuture of mouse pacsin3 bar domain mutant
69 c3pdgA_
not modelled
15.1
18
PDB header: unknown functionChain: A: PDB Molecule: fibronectin(iii)-like module;PDBTitle: structures of clostridium thermocellum cbha fibronectin(iii)-like2 modules
70 c1gk4A_
not modelled
15.1
10
PDB header: vimentinChain: A: PDB Molecule: vimentin;PDBTitle: human vimentin coil 2b fragment (cys2)
71 d1ji5a_
not modelled
14.8
16
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin72 d1u5pa2
not modelled
14.7
20
Fold: Spectrin repeat-likeSuperfamily: Spectrin repeatFamily: Spectrin repeat73 c2pc6C_
not modelled
14.7
19
PDB header: lyaseChain: C: PDB Molecule: probable acetolactate synthase isozyme iii (small subunit);PDBTitle: crystal structure of putative acetolactate synthase- small subunit2 from nitrosomonas europaea
74 d1ru3a_
not modelled
14.6
19
Fold: Prismane protein-likeSuperfamily: Prismane protein-likeFamily: Acetyl-CoA synthase75 d2fgca2
not modelled
14.6
28
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: IlvH-like76 d1p7hl1
not modelled
14.5
13
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: NF-kappa-B/REL/DORSAL transcription factors, C-terminal domain77 c2k6lA_
not modelled
14.5
44
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: the solution structure of xacb0070 from xanthonomas2 axonopodis pv citri reveals this new protein is a member of3 the rhh family of transcriptional repressors
78 d1nj8a3
not modelled
14.5
18
Fold: Class II aaRS and biotin synthetasesSuperfamily: Class II aaRS and biotin synthetasesFamily: Class II aminoacyl-tRNA synthetase (aaRS)-like, catalytic domain79 c3bbnI_
not modelled
14.4
19
PDB header: ribosomeChain: I: PDB Molecule: ribosomal protein s9;PDBTitle: homology model for the spinach chloroplast 30s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome.
80 c2c6rA_
not modelled
14.3
14
PDB header: dna-binding proteinChain: A: PDB Molecule: dna-binding stress response protein, dps family;PDBTitle: fe-soaked crystal structure of the dps92 from deinococcus2 radiodurans
81 c3bboH_
not modelled
14.0
26
PDB header: ribosomeChain: H: PDB Molecule: ribosomal protein l5;PDBTitle: homology model for the spinach chloroplast 50s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome
82 c2z9fC_
not modelled
13.9
37
PDB header: biosynthetic proteinChain: C: PDB Molecule: cellulose synthase operon protein d;PDBTitle: crystal structure of axcesd protein from acetobacter xylinum
83 d2j8wa1
not modelled
13.6
18
Fold: Four-helical up-and-down bundleSuperfamily: CytochromesFamily: Cytochrome c'-like84 c2xzmI_
not modelled
13.5
23
PDB header: ribosomeChain: I: PDB Molecule: rps16e;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
85 d2f1fa1
not modelled
13.1
18
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: IlvH-like86 d1oaoc_
not modelled
13.1
19
Fold: Prismane protein-likeSuperfamily: Prismane protein-likeFamily: Acetyl-CoA synthase87 c2f7nA_
not modelled
13.0
17
PDB header: dna binding proteinChain: A: PDB Molecule: dna-binding stress response protein, dps family;PDBTitle: structure of d. radiodurans dps-1
88 c2zkqi_
not modelled
12.8
29
PDB header: ribosomal protein/rnaChain: I: PDB Molecule: PDBTitle: structure of a mammalian ribosomal 40s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
89 d2f66c1
not modelled
12.7
47
Fold: Long alpha-hairpinSuperfamily: Endosomal sorting complex assembly domainFamily: VPS37 C-terminal domain-like90 d2gu3a1
not modelled
12.1
21
Fold: Cystatin-likeSuperfamily: Cystatin/monellinFamily: PepSY-like91 c2lf0A_
not modelled
12.0
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein yibl;PDBTitle: solution structure of sf3636, a two-domain unknown function protein2 from shigella flexneri 2a, determined by joint refinement of nmr,3 residual dipolar couplings and small-angle x-ray scatting, nesg4 target sfr339/ocsp target sf3636
92 d1utya_
not modelled
11.8
15
Fold: BTV NS2-like ssRNA-binding domainSuperfamily: BTV NS2-like ssRNA-binding domainFamily: BTV NS2-like ssRNA-binding domain93 c1utyA_
not modelled
11.8
15
PDB header: viral proteinChain: A: PDB Molecule: non-structural protein 2;PDBTitle: crystal structure of the rna binding domain of bluetongue2 virus non-structural protein 2(ns2)
94 c2jzxA_
not modelled
11.4
21
PDB header: rna binding proteinChain: A: PDB Molecule: poly(rc)-binding protein 2;PDBTitle: pcbp2 kh1-kh2 domains
95 d1u5pa1
not modelled
11.4
16
Fold: Spectrin repeat-likeSuperfamily: Spectrin repeatFamily: Spectrin repeat96 c3pdyB_
not modelled
11.2
11
PDB header: structural proteinChain: B: PDB Molecule: plectin;PDBTitle: structure of the third and fourth spectrin repeats of the plakin2 domain of plectin
97 d2g5ca1
not modelled
11.1
19
Fold: 6-phosphogluconate dehydrogenase C-terminal domain-likeSuperfamily: 6-phosphogluconate dehydrogenase C-terminal domain-likeFamily: TyrA dimerization domain-like98 d1e85a_
not modelled
11.1
7
Fold: Four-helical up-and-down bundleSuperfamily: CytochromesFamily: Cytochrome c'-like99 c2obaE_
not modelled
10.9
24
PDB header: lyaseChain: E: PDB Molecule: probable 6-pyruvoyl tetrahydrobiopterin synthase;PDBTitle: pseudomonas aeruginosa 6-pyruvoyl tetrahydrobiopterin synthase