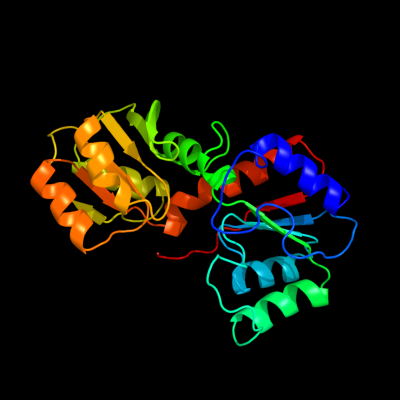
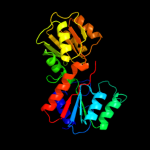
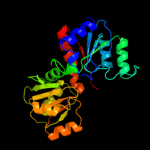
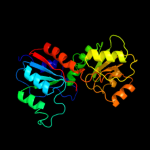
1 d1nmpa_
100.0
100
Fold: NIF3 (NGG1p interacting factor 3)-likeSuperfamily: NIF3 (NGG1p interacting factor 3)-likeFamily: NIF3 (NGG1p interacting factor 3)-like2 c2nydB_
100.0
23
PDB header: unknown functionChain: B: PDB Molecule: upf0135 protein sa1388;PDBTitle: crystal structure of staphylococcus aureus hypothetical protein sa1388
3 c2gx8B_
100.0
25
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: nif3-related protein;PDBTitle: the crystal stucture of bacillus cereus protein related to nif3
4 d2gx8a1
100.0
25
Fold: NIF3 (NGG1p interacting factor 3)-likeSuperfamily: NIF3 (NGG1p interacting factor 3)-likeFamily: NIF3 (NGG1p interacting factor 3)-like5 c2yybA_
100.0
37
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein ttha1606;PDBTitle: crystal structure of ttha1606 from thermus thermophilus hb8
6 d2fywa1
100.0
24
Fold: NIF3 (NGG1p interacting factor 3)-likeSuperfamily: NIF3 (NGG1p interacting factor 3)-likeFamily: NIF3 (NGG1p interacting factor 3)-like7 c3rxyA_
99.9
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: nif3 protein;PDBTitle: crystal structure of nif3 superfamily protein from sphaerobacter2 thermophilus
8 c2yx6C_
85.0
12
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: hypothetical protein ph0822;PDBTitle: crystal structure of ph0822
9 c3qc3B_
83.4
14
PDB header: isomeraseChain: B: PDB Molecule: d-ribulose-5-phosphate-3-epimerase;PDBTitle: crystal structure of a d-ribulose-5-phosphate-3-epimerase (np_954699)2 from homo sapiens at 2.20 a resolution
10 c2am1A_
79.3
11
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramoylalanine-d-glutamyl-lysine-d-alanyl-d-PDBTitle: sp protein ligand 1
11 d1w0ma_
77.7
18
Fold: TIM beta/alpha-barrelSuperfamily: Triosephosphate isomerase (TIM)Family: Triosephosphate isomerase (TIM)12 d1t3va_
75.8
13
Fold: Ribonuclease H-like motifSuperfamily: Nitrogenase accessory factor-likeFamily: MTH1175-like13 d1eo1a_
75.4
23
Fold: Ribonuclease H-like motifSuperfamily: Nitrogenase accessory factor-likeFamily: MTH1175-like14 c3e96B_
70.8
19
PDB header: lyaseChain: B: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from2 bacillus clausii
15 c2wfbA_
67.3
20
PDB header: biosynthetic proteinChain: A: PDB Molecule: putative uncharacterized protein orp;PDBTitle: high resolution structure of the apo form of the orange2 protein (orp) from desulfovibrio gigas
16 c3d0cB_
66.0
22
PDB header: lyaseChain: B: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from2 oceanobacillus iheyensis at 1.9 a resolution
17 c3aamA_
65.6
15
PDB header: hydrolaseChain: A: PDB Molecule: endonuclease iv;PDBTitle: crystal structure of endonuclease iv from thermus thermophilus hb8
18 c3c6kC_
64.4
14
PDB header: transferaseChain: C: PDB Molecule: spermine synthase;PDBTitle: crystal structure of human spermine synthase in complex2 with spermidine and 5-methylthioadenosine
19 c3ct7E_
64.1
17
PDB header: isomeraseChain: E: PDB Molecule: d-allulose-6-phosphate 3-epimerase;PDBTitle: crystal structure of d-allulose 6-phosphate 3-epimerase2 from escherichia coli k-12
20 d1p3da1
63.9
17
Fold: MurCD N-terminal domainSuperfamily: MurCD N-terminal domainFamily: MurCD N-terminal domain21 c3jr2D_
not modelled
61.3
14
PDB header: biosynthetic proteinChain: D: PDB Molecule: hexulose-6-phosphate synthase sgbh;PDBTitle: x-ray crystal structure of the mg-bound 3-keto-l-gulonate-6-phosphate2 decarboxylase from vibrio cholerae o1 biovar el tor str. n16961
22 d1afsa_
not modelled
59.8
9
Fold: TIM beta/alpha-barrelSuperfamily: NAD(P)-linked oxidoreductaseFamily: Aldo-keto reductases (NADP)23 c2yw3E_
not modelled
59.3
16
PDB header: lyaseChain: E: PDB Molecule: 4-hydroxy-2-oxoglutarate aldolase/2-deydro-3-PDBTitle: crystal structure analysis of the 4-hydroxy-2-oxoglutarate aldolase/2-2 deydro-3-deoxyphosphogluconate aldolase from tthb1
24 d1u83a_
not modelled
58.1
16
Fold: TIM beta/alpha-barrelSuperfamily: (2r)-phospho-3-sulfolactate synthase ComAFamily: (2r)-phospho-3-sulfolactate synthase ComA25 c1u83A_
not modelled
58.1
16
PDB header: lyaseChain: A: PDB Molecule: phosphosulfolactate synthase;PDBTitle: psl synthase from bacillus subtilis
26 d1us0a_
not modelled
57.6
13
Fold: TIM beta/alpha-barrelSuperfamily: NAD(P)-linked oxidoreductaseFamily: Aldo-keto reductases (NADP)27 d1xp3a1
not modelled
55.8
22
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Endonuclease IV28 d1e7wa_
not modelled
55.5
24
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases29 d1rdua_
not modelled
52.5
18
Fold: Ribonuclease H-like motifSuperfamily: Nitrogenase accessory factor-likeFamily: MTH1175-like30 d2flia1
not modelled
52.1
16
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: D-ribulose-5-phosphate 3-epimerase31 c3o26A_
not modelled
51.5
32
PDB header: oxidoreductaseChain: A: PDB Molecule: salutaridine reductase;PDBTitle: the structure of salutaridine reductase from papaver somniferum.
32 c3p19A_
not modelled
51.0
18
PDB header: oxidoreductaseChain: A: PDB Molecule: putative blue fluorescent protein;PDBTitle: improved nadph-dependent blue fluorescent protein
33 d1p90a_
not modelled
50.5
6
Fold: Ribonuclease H-like motifSuperfamily: Nitrogenase accessory factor-likeFamily: Nitrogenase accessory factor34 d3clsc1
not modelled
47.9
16
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: ETFP subunits35 c2re2A_
not modelled
47.9
12
PDB header: oxidoreductaseChain: A: PDB Molecule: uncharacterized protein ta1041;PDBTitle: crystal structure of a putative iron-molybdenum cofactor (femo-co)2 dinitrogenase (ta1041m) from thermoplasma acidophilum dsm 1728 at3 1.30 a resolution
36 d1p33a_
not modelled
47.1
20
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases37 d1h1ya_
not modelled
46.6
16
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: D-ribulose-5-phosphate 3-epimerase38 c3pdiB_
not modelled
46.2
17
PDB header: protein bindingChain: B: PDB Molecule: nitrogenase mofe cofactor biosynthesis protein nifn;PDBTitle: precursor bound nifen
39 d1j6ua1
not modelled
46.1
14
Fold: MurCD N-terminal domainSuperfamily: MurCD N-terminal domainFamily: MurCD N-terminal domain40 c2foiB_
not modelled
44.6
18
PDB header: oxidoreductaseChain: B: PDB Molecule: enoyl-acyl carrier reductase;PDBTitle: synthesis, biological activity, and x-ray crystal structural analysis2 of diaryl ether inhibitors of malarial enoyl acp reductase.
41 c2nq8B_
not modelled
44.6
18
PDB header: oxidoreductaseChain: B: PDB Molecule: enoyl-acyl carrier reductase;PDBTitle: malarial enoyl acyl acp reductase bound with inh-nad adduct
42 c2wtzC_
not modelled
44.6
14
PDB header: ligaseChain: C: PDB Molecule: udp-n-acetylmuramoyl-l-alanyl-d-glutamate-PDBTitle: mure ligase of mycobacterium tuberculosis
43 c3ppiA_
not modelled
43.9
22
PDB header: oxidoreductaseChain: A: PDB Molecule: 3-hydroxyacyl-coa dehydrogenase type-2;PDBTitle: crystal structure of 3-hydroxyacyl-coa dehydrogenase type-2 from2 mycobacterium avium
44 d2pv7a2
not modelled
42.7
17
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain45 d1uh5a_
not modelled
41.9
18
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases46 d1km4a_
not modelled
41.7
17
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Decarboxylase47 d1rpxa_
not modelled
40.9
15
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: D-ribulose-5-phosphate 3-epimerase48 c1zcjA_
not modelled
40.7
17
PDB header: oxidoreductaseChain: A: PDB Molecule: peroxisomal bifunctional enzyme;PDBTitle: crystal structure of 3-hydroxyacyl-coa dehydrogenase
49 c3lf2B_
not modelled
40.3
30
PDB header: oxidoreductaseChain: B: PDB Molecule: short chain oxidoreductase q9hya2;PDBTitle: nadph bound structure of the short chain oxidoreductase q9hya2 from2 pseudomonas aeruginosa pao1 containing an atypical catalytic center
50 d1o13a_
not modelled
39.6
7
Fold: Ribonuclease H-like motifSuperfamily: Nitrogenase accessory factor-likeFamily: MTH1175-like51 c3inpA_
not modelled
39.5
20
PDB header: isomeraseChain: A: PDB Molecule: d-ribulose-phosphate 3-epimerase;PDBTitle: 2.05 angstrom resolution crystal structure of d-ribulose-phosphate 3-2 epimerase from francisella tularensis.
52 c3t4xA_
not modelled
39.0
33
PDB header: oxidoreductaseChain: A: PDB Molecule: oxidoreductase, short chain dehydrogenase/reductase family;PDBTitle: short chain dehydrogenase/reductase family oxidoreductase from2 bacillus anthracis str. ames ancestor
53 d1gega_
not modelled
38.8
20
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases54 d1yxma1
not modelled
38.7
33
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases55 c3l77A_
not modelled
38.6
33
PDB header: oxidoreductaseChain: A: PDB Molecule: short-chain alcohol dehydrogenase;PDBTitle: x-ray structure alcohol dehydrogenase from archaeon thermococcus2 sibiricus complexed with 5-hydroxy-nadp
56 c3dhnA_
not modelled
38.2
17
PDB header: isomerase, lyaseChain: A: PDB Molecule: nad-dependent epimerase/dehydratase;PDBTitle: crystal structure of the putative epimerase q89z24_bactn2 from bacteroides thetaiotaomicron. northeast structural3 genomics consortium target btr310.
57 c3noeA_
not modelled
38.1
17
PDB header: lyaseChain: A: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from pseudomonas2 aeruginosa
58 c2ehhE_
not modelled
37.8
14
PDB header: lyaseChain: E: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from2 aquifex aeolicus
59 d2hmva1
not modelled
37.7
17
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Potassium channel NAD-binding domain60 d1nu0a_
not modelled
36.3
13
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Putative Holliday junction resolvase RuvX61 d1fjha_
not modelled
36.3
15
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases62 d1dvja_
not modelled
36.0
16
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Decarboxylase63 c2ntnB_
not modelled
35.3
33
PDB header: oxidoreductaseChain: B: PDB Molecule: 3-oxoacyl-[acyl-carrier-protein] reductase;PDBTitle: crystal structure of maba-c60v/g139a/s144l
64 c3s55F_
not modelled
34.9
41
PDB header: oxidoreductaseChain: F: PDB Molecule: putative short-chain dehydrogenase/reductase;PDBTitle: crystal structure of a putative short-chain dehydrogenase/reductase2 from mycobacterium abscessus bound to nad
65 d2gv8a1
not modelled
34.2
13
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains66 d1uaya_
not modelled
34.1
30
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases67 c3bmrA_
not modelled
33.7
22
PDB header: oxidoreductaseChain: A: PDB Molecule: pteridine reductase;PDBTitle: structure of pteridine reductase 1 (ptr1) from trypanosoma2 brucei in ternary complex with cofactor (nadp+) and3 inhibitor (compound ax6)
68 c3un1D_
not modelled
33.5
25
PDB header: oxidoreductaseChain: D: PDB Molecule: probable oxidoreductase;PDBTitle: crystal structure of an oxidoreductase from sinorhizobium meliloti2 1021
69 d1vhxa_
not modelled
33.3
13
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Putative Holliday junction resolvase RuvX70 c2x58B_
not modelled
33.2
17
PDB header: oxidoreductaseChain: B: PDB Molecule: peroxisomal bifunctional enzyme;PDBTitle: the crystal structure of mfe1 liganded with coa
71 c2vdcI_
not modelled
32.9
12
PDB header: oxidoreductaseChain: I: PDB Molecule: glutamate synthase [nadph] small chain;PDBTitle: the 9.5 a resolution structure of glutamate synthase from2 cryo-electron microscopy and its oligomerization behavior3 in solution: functional implications.
72 d2gdza1
not modelled
32.8
30
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases73 c3gk3D_
not modelled
32.5
20
PDB header: oxidoreductaseChain: D: PDB Molecule: acetoacetyl-coa reductase;PDBTitle: crystal structure of acetoacetyl-coa reductase from2 burkholderia pseudomallei 1710b
74 c3g0sA_
not modelled
32.5
12
PDB header: lyaseChain: A: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: dihydrodipicolinate synthase from salmonella typhimurium lt2
75 c3tfoD_
not modelled
31.9
26
PDB header: oxidoreductaseChain: D: PDB Molecule: putative 3-oxoacyl-(acyl-carrier-protein) reductase;PDBTitle: crystal structure of a putative 3-oxoacyl-(acyl-carrier-protein)2 reductase from sinorhizobium meliloti
76 c3dz1A_
not modelled
31.7
17
PDB header: lyaseChain: A: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 1.87a resolution
77 c2p91A_
not modelled
31.5
26
PDB header: oxidoreductaseChain: A: PDB Molecule: enoyl-[acyl-carrier-protein] reductase [nadh];PDBTitle: crystal structure of enoyl-[acyl-carrier-protein] reductase (nadh)2 from aquifex aeolicus vf5
78 c2nm0B_
not modelled
31.5
36
PDB header: oxidoreductaseChain: B: PDB Molecule: probable 3-oxacyl-(acyl-carrier-protein) reductase;PDBTitle: crystal structure of sco1815: a beta-ketoacyl-acyl carrier protein2 reductase from streptomyces coelicolor a3(2)
79 d1xq1a_
not modelled
31.5
30
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases80 c3k6jA_
not modelled
31.2
24
PDB header: oxidoreductaseChain: A: PDB Molecule: protein f01g10.3, confirmed by transcript evidence;PDBTitle: crystal structure of the dehydrogenase part of multifuctional enzyme 12 from c.elegans
81 d2d1ya1
not modelled
30.8
30
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases82 c2pv7B_
not modelled
30.6
17
PDB header: isomerase, oxidoreductaseChain: B: PDB Molecule: t-protein [includes: chorismate mutase (ec 5.4.99.5) (cm)PDBTitle: crystal structure of chorismate mutase / prephenate dehydrogenase2 (tyra) (1574749) from haemophilus influenzae rd at 2.00 a resolution
83 d1xg5a_
not modelled
30.2
30
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases84 c2qhxB_
not modelled
30.0
28
PDB header: oxidoreductaseChain: B: PDB Molecule: pteridine reductase 1;PDBTitle: structure of pteridine reductase from leishmania major2 complexed with a ligand
85 c3i1jB_
not modelled
29.9
33
PDB header: oxidoreductaseChain: B: PDB Molecule: oxidoreductase, short chainPDBTitle: structure of a putative short chain dehydrogenase from2 pseudomonas syringae
86 d1f6ya_
not modelled
29.4
15
Fold: TIM beta/alpha-barrelSuperfamily: Dihydropteroate synthetase-likeFamily: Methyltetrahydrofolate-utiluzing methyltransferases87 c2vt2A_
not modelled
29.1
18
PDB header: transcriptionChain: A: PDB Molecule: redox-sensing transcriptional repressor rex;PDBTitle: structure and functional properties of the bacillus2 subtilis transcriptional repressor rex
88 c3rihB_
not modelled
28.7
33
PDB header: oxidoreductaseChain: B: PDB Molecule: short chain dehydrogenase or reductase;PDBTitle: crystal structure of a putative short chain dehydrogenase or reductase2 from mycobacterium abscessus
89 d1ah4a_
not modelled
28.6
13
Fold: TIM beta/alpha-barrelSuperfamily: NAD(P)-linked oxidoreductaseFamily: Aldo-keto reductases (NADP)90 c3f5sA_
not modelled
28.5
33
PDB header: oxidoreductaseChain: A: PDB Molecule: dehydrogenase;PDBTitle: crystal structure of putatitve short chain dehydrogenase from shigella2 flexneri 2a str. 301
91 c3n2xB_
not modelled
28.4
18
PDB header: lyaseChain: B: PDB Molecule: uncharacterized protein yage;PDBTitle: crystal structure of yage, a prophage protein belonging to the2 dihydrodipicolinic acid synthase family from e. coli k12 in complex3 with pyruvate
92 c3uxyC_
not modelled
28.1
30
PDB header: oxidoreductaseChain: C: PDB Molecule: short-chain dehydrogenase/reductase sdr;PDBTitle: the crystal structure of short chain dehydrogenase from rhodobacter2 sphaeroides
93 c3n0vD_
not modelled
28.1
13
PDB header: hydrolaseChain: D: PDB Molecule: formyltetrahydrofolate deformylase;PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (pp_0327)2 from pseudomonas putida kt2440 at 2.25 a resolution
94 d2h7ma1
not modelled
28.1
19
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases95 d1zk4a1
not modelled
28.0
41
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases96 c1zmoF_
not modelled
28.0
15
PDB header: lyaseChain: F: PDB Molecule: halohydrin dehalogenase;PDBTitle: apo structure of haloalcohol dehalogenase hhea of2 arthrobacter sp. ad2
97 d2bela_
not modelled
27.8
26
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases98 d1vqta1
not modelled
27.4
13
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Decarboxylase99 d1s1pa_
not modelled
27.3
11
Fold: TIM beta/alpha-barrelSuperfamily: NAD(P)-linked oxidoreductaseFamily: Aldo-keto reductases (NADP)100 c3louB_
not modelled
27.2
12
PDB header: hydrolaseChain: B: PDB Molecule: formyltetrahydrofolate deformylase;PDBTitle: crystal structure of formyltetrahydrofolate deformylase (yp_105254.1)2 from burkholderia mallei atcc 23344 at 1.90 a resolution
101 c3sc4A_
not modelled
27.0
22
PDB header: oxidoreductaseChain: A: PDB Molecule: short chain dehydrogenase (a0qtm2 homolog);PDBTitle: crystal structure of a short chain dehydrogenase (a0qtm2 homolog)2 mycobacterium thermoresistibile
102 c3qivA_
not modelled
26.8
41
PDB header: oxidoreductaseChain: A: PDB Molecule: short-chain dehydrogenase or 3-oxoacyl-[acyl-carrier-PDBTitle: crystal structure of a putative short-chain dehydrogenase or 3-2 oxoacyl-[acyl-carrier-protein] reductase from mycobacterium3 paratuberculosis atcc baa-968 / k-10
103 c2ydyA_
not modelled
26.7
21
PDB header: oxidoreductaseChain: A: PDB Molecule: methionine adenosyltransferase 2 subunit beta;PDBTitle: crystal structure of human s-adenosylmethionine synthetase2 2, beta subunit in orthorhombic crystal form
104 d1vjda_
not modelled
26.7
15
Fold: Phosphoglycerate kinaseSuperfamily: Phosphoglycerate kinaseFamily: Phosphoglycerate kinase105 c2dknA_
not modelled
26.4
30
PDB header: oxidoreductaseChain: A: PDB Molecule: 3-alpha-hydroxysteroid dehydrogenase;PDBTitle: crystal structure of the 3-alpha-hydroxysteroid dehydrogenase from2 pseudomonas sp. b-0831 complexed with nadh
106 c2w1vA_
not modelled
26.1
7
PDB header: hydrolaseChain: A: PDB Molecule: nitrilase homolog 2;PDBTitle: crystal structure of mouse nitrilase-2 at 1.4a resolution
107 d1xcfa_
not modelled
26.1
17
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes108 c3ezlA_
not modelled
26.0
20
PDB header: oxidoreductaseChain: A: PDB Molecule: acetoacetyl-coa reductase;PDBTitle: crystal structure of acetyacetyl-coa reductase from2 burkholderia pseudomallei 1710b
109 c1yb1B_
not modelled
25.8
22
PDB header: oxidoreductaseChain: B: PDB Molecule: 17-beta-hydroxysteroid dehydrogenase type xi;PDBTitle: crystal structure of human 17-beta-hydroxysteroid dehydrogenase type2 xi
110 d1xu9a_
not modelled
25.8
14
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases111 c2x7vA_
not modelled
25.7
11
PDB header: hydrolaseChain: A: PDB Molecule: probable endonuclease 4;PDBTitle: crystal structure of thermotoga maritima endonuclease iv in2 the presence of zinc
112 c3tfxB_
not modelled
25.4
19
PDB header: lyaseChain: B: PDB Molecule: orotidine 5'-phosphate decarboxylase;PDBTitle: crystal structure of orotidine 5'-phosphate decarboxylase from2 lactobacillus acidophilus
113 d1o94c_
not modelled
25.3
17
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: ETFP subunits114 d1gtea3
not modelled
25.3
16
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: C-terminal domain of adrenodoxin reductase-like115 c3rd5A_
not modelled
25.1
30
PDB header: oxidoreductaseChain: A: PDB Molecule: mypaa.01249.c;PDBTitle: crystal structure of a putative uncharacterized protein from2 mycobacterium paratuberculosis
116 c3q2oB_
not modelled
24.8
18
PDB header: lyaseChain: B: PDB Molecule: phosphoribosylaminoimidazole carboxylase, atpase subunit;PDBTitle: crystal structure of purk: n5-carboxyaminoimidazole ribonucleotide2 synthetase
117 d1rd5a_
not modelled
24.7
20
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes118 d1x1ta1
not modelled
24.6
37
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases119 c3uvzB_
not modelled
24.4
22
PDB header: lyaseChain: B: PDB Molecule: phosphoribosylaminoimidazole carboxylase, atpase subunit;PDBTitle: crystal structure of phosphoribosylaminoimidazole carboxylase, atpase2 subunit from burkholderia ambifaria
120 c2hjwA_
not modelled
24.3
16
PDB header: ligaseChain: A: PDB Molecule: acetyl-coa carboxylase 2;PDBTitle: crystal structure of the bc domain of acc2