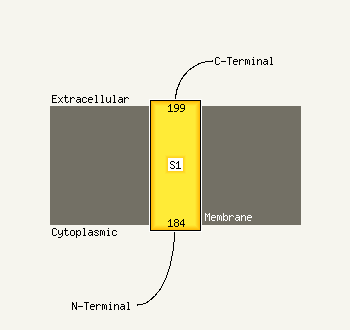
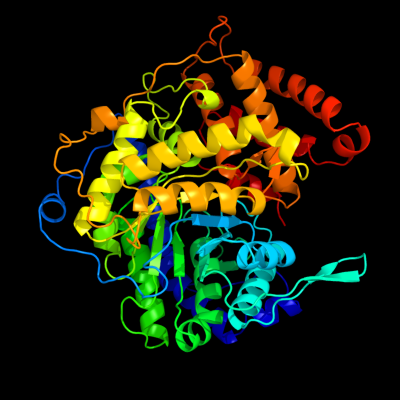
| 1 | c3cuzA_
|
|
 |
100.0 |
100 |
PDB header:transferase
Chain: A: PDB Molecule:malate synthase a;
PDBTitle: atomic resolution structures of escherichia coli and2 bacillis anthracis malate synthase a: comparison with3 isoform g and implications for structure based drug design
|
| 2 | c3cuxA_
|
|
 |
100.0 |
55 |
PDB header:transferase
Chain: A: PDB Molecule:malate synthase;
PDBTitle: atomic resolution structures of escherichia coli and2 bacillis anthracis malate synthase a: comparison with3 isoform g and implications for structure based drug design
|
| 3 | d1n8ia_
|
|
 |
100.0 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Malate synthase G
Family:Malate synthase G |
| 4 | d1d8ca_
|
|
 |
100.0 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Malate synthase G
Family:Malate synthase G |
| 5 | c3pugA_
|
|
 |
100.0 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:malate synthase;
PDBTitle: haloferax volcanii malate synthase native at 3mm glyoxylate
|
| 6 | c3r4iB_
|
|
 |
100.0 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:citrate lyase;
PDBTitle: crystal structure of a citrate lyase (bxe_b2899) from burkholderia2 xenovorans lb400 at 2.24 a resolution
|
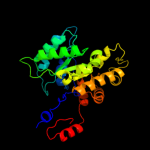
| 7 | c3qqwC_
|
|
 |
100.0 |
15 |
PDB header:lyase
Chain: C: PDB Molecule:putative citrate lyase;
PDBTitle: crystal structure of a hypothetical lyase (reut_b4148) from ralstonia2 eutropha jmp134 at 2.44 a resolution
|
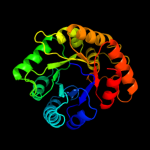
| 8 | c1sgjB_
|
|
 |
100.0 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:citrate lyase, beta subunit;
PDBTitle: crystal structure of citrate lyase beta subunit
|
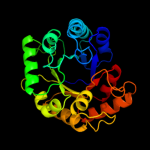
| 9 | d1sgja_
|
|
 |
100.0 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:HpcH/HpaI aldolase |
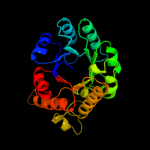
| 10 | c1u5vA_
|
|
 |
100.0 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:cite;
PDBTitle: structure of cite complexed with triphosphate group of atp2 form mycobacterium tuberculosis
|
| 11 | d1u5ha_
|
|
 |
100.0 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:HpcH/HpaI aldolase |
| 12 | d1dxea_
|
|
 |
99.9 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:HpcH/HpaI aldolase |
| 13 | c2v5jB_
|
|
 |
99.8 |
20 |
PDB header:lyase
Chain: B: PDB Molecule:2,4-dihydroxyhept-2-ene-1,7-dioic acid aldolase;
PDBTitle: apo class ii aldolase hpch
|
| 14 | c1izcA_
|
|
 |
99.8 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:macrophomate synthase intermolecular diels-alderase;
PDBTitle: crystal structure analysis of macrophomate synthase
|
| 15 | d1izca_
|
|
 |
99.8 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:HpcH/HpaI aldolase |
| 16 | c2vwtA_
|
|
 |
99.8 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:yfau, 2-keto-3-deoxy sugar aldolase;
PDBTitle: crystal structure of yfau, a metal ion dependent class ii2 aldolase from escherichia coli k12 - mg-pyruvate product3 complex
|
| 17 | c3qz6A_
|
|
 |
99.7 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:hpch/hpai aldolase;
PDBTitle: the crystal structure of hpch/hpai aldolase from desulfitobacterium2 hafniense dcb-2
|
| 18 | d1e0ta2
|
|
 |
99.7 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 19 | d1a3xa2
|
|
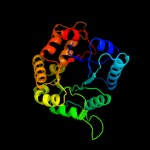 |
99.6 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 20 | d1pkla2
|
|
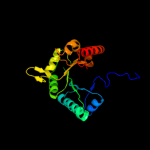 |
99.4 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 21 | d2g50a2 |
|
not modelled |
99.3 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 22 | d1liua2 |
|
not modelled |
99.3 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 23 | c2hwgA_ |
|
not modelled |
98.9 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoenolpyruvate-protein phosphotransferase;
PDBTitle: structure of phosphorylated enzyme i of the2 phosphoenolpyruvate:sugar phosphotransferase system
|
| 24 | c2hroA_ |
|
not modelled |
98.8 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoenolpyruvate-protein phosphotransferase;
PDBTitle: structure of the full-lenght enzyme i of the pts system from2 staphylococcus carnosus
|
| 25 | c2bg5C_ |
|
not modelled |
98.8 |
14 |
PDB header:transferase
Chain: C: PDB Molecule:phosphoenolpyruvate-protein kinase;
PDBTitle: crystal structure of the phosphoenolpyruvate-binding enzyme2 i-domain from the thermoanaerobacter tengcongensis pep:3 sugar phosphotransferase system (pts)
|
| 26 | d1h6za1 |
|
not modelled |
98.7 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate phosphate dikinase, C-terminal domain |
| 27 | d1vbga1 |
|
not modelled |
98.6 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate phosphate dikinase, C-terminal domain |
| 28 | c1vbhA_ |
|
not modelled |
98.6 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:pyruvate,orthophosphate dikinase;
PDBTitle: pyruvate phosphate dikinase with bound mg-pep from maize
|
| 29 | c1t5aB_ |
|
not modelled |
98.4 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:pyruvate kinase, m2 isozyme;
PDBTitle: human pyruvate kinase m2
|
| 30 | c2vgbB_ |
|
not modelled |
98.3 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:pyruvate kinase isozymes r/l;
PDBTitle: human erythrocyte pyruvate kinase
|
| 31 | c1aqfB_ |
|
not modelled |
98.3 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:pyruvate kinase;
PDBTitle: pyruvate kinase from rabbit muscle with mg, k, and l-2 phospholactate
|
| 32 | c3khdC_ |
|
not modelled |
98.3 |
20 |
PDB header:transferase
Chain: C: PDB Molecule:pyruvate kinase;
PDBTitle: crystal structure of pff1300w.
|
| 33 | c1h6zA_ |
|
not modelled |
98.3 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:pyruvate phosphate dikinase;
PDBTitle: 3.0 a resolution crystal structure of glycosomal pyruvate2 phosphate dikinase from trypanosoma brucei
|
| 34 | d1kbla1 |
|
not modelled |
98.1 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate phosphate dikinase, C-terminal domain |
| 35 | c1e0tD_ |
|
not modelled |
98.1 |
19 |
PDB header:phosphotransferase
Chain: D: PDB Molecule:pyruvate kinase;
PDBTitle: r292d mutant of e. coli pyruvate kinase
|
| 36 | c1kblA_ |
|
not modelled |
98.1 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:pyruvate phosphate dikinase;
PDBTitle: pyruvate phosphate dikinase
|
| 37 | c1a3wB_ |
|
not modelled |
98.0 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:pyruvate kinase;
PDBTitle: pyruvate kinase from saccharomyces cerevisiae complexed with fbp, pg,2 mn2+ and k+
|
| 38 | c3ma8A_ |
|
not modelled |
98.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:pyruvate kinase;
PDBTitle: crystal structure of cgd1_2040, a pyruvate kinase from cryptosporidium2 parvum
|
| 39 | c1pklB_ |
|
not modelled |
97.9 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:protein (pyruvate kinase);
PDBTitle: the structure of leishmania pyruvate kinase
|
| 40 | c2olsA_ |
|
not modelled |
97.9 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoenolpyruvate synthase;
PDBTitle: the crystal structure of the phosphoenolpyruvate synthase from2 neisseria meningitidis
|
| 41 | c3e0vB_ |
|
not modelled |
97.8 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:pyruvate kinase;
PDBTitle: crystal structure of pyruvate kinase from leishmania mexicana in2 complex with sulphate ions
|
| 42 | c2e28A_ |
|
not modelled |
97.7 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:pyruvate kinase;
PDBTitle: crystal structure analysis of pyruvate kinase from bacillus2 stearothermophilus
|
| 43 | c3eoeC_ |
|
not modelled |
97.7 |
18 |
PDB header:transferase
Chain: C: PDB Molecule:pyruvate kinase;
PDBTitle: crystal structure of pyruvate kinase from toxoplasma gondii, 55.m00007
|
| 44 | c3t07D_ |
|
not modelled |
97.6 |
20 |
PDB header:transferase/transferase inhibitor
Chain: D: PDB Molecule:pyruvate kinase;
PDBTitle: crystal structure of s. aureus pyruvate kinase in complex with a2 naturally occurring bis-indole alkaloid
|
| 45 | c3odmE_ |
|
not modelled |
97.6 |
19 |
PDB header:lyase
Chain: E: PDB Molecule:phosphoenolpyruvate carboxylase;
PDBTitle: archaeal-type phosphoenolpyruvate carboxylase
|
| 46 | d1jqoa_ |
|
not modelled |
76.0 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate carboxylase |
| 47 | c1jqoA_ |
|
not modelled |
76.0 |
10 |
PDB header:lyase
Chain: A: PDB Molecule:phosphoenolpyruvate carboxylase;
PDBTitle: crystal structure of c4-form phosphoenolpyruvate carboxylase from2 maize
|
| 48 | d1jqna_ |
|
not modelled |
71.4 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate carboxylase |
| 49 | c3r3sD_ |
|
not modelled |
62.3 |
17 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:oxidoreductase;
PDBTitle: structure of the ygha oxidoreductase from salmonella enterica
|
| 50 | c3i1jB_ |
|
not modelled |
61.0 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase, short chain
PDBTitle: structure of a putative short chain dehydrogenase from2 pseudomonas syringae
|
| 51 | c2x3mA_ |
|
not modelled |
52.0 |
36 |
PDB header:unknown function
Chain: A: PDB Molecule:hypothetical protein orf239;
PDBTitle: crystal structure of hypothetical protein orf239 from pyrobaculum2 spherical virus
|
| 52 | c3g5oC_ |
|
not modelled |
40.9 |
13 |
PDB header:toxin/antitoxin
Chain: C: PDB Molecule:uncharacterized protein rv2866;
PDBTitle: the crystal structure of the toxin-antitoxin complex relbe2 (rv2865-2 2866) from mycobacterium tuberculosis
|
| 53 | d1yxma1 |
|
not modelled |
40.9 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 54 | d1qasa3 |
|
not modelled |
35.5 |
28 |
Fold:TIM beta/alpha-barrel
Superfamily:PLC-like phosphodiesterases
Family:Mammalian PLC |
| 55 | d1g0oa_ |
|
not modelled |
30.2 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 56 | d2zkmx4 |
|
not modelled |
28.4 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:PLC-like phosphodiesterases
Family:Mammalian PLC |
| 57 | c3iv6C_ |
|
not modelled |
26.6 |
29 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative zn-dependent alcohol dehydrogenase;
PDBTitle: crystal structure of putative zn-dependent alcohol dehydrogenases from2 rhodobacter sphaeroides.
|
| 58 | c2gjmA_ |
|
not modelled |
26.1 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:lactoperoxidase;
PDBTitle: crystal structure of buffalo lactoperoxidase at 2.75a resolution
|
| 59 | c3qkbB_ |
|
not modelled |
25.4 |
10 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a protein with unknown function which belongs to2 pfam duf74 family (pepe_0654) from pediococcus pentosaceus atcc 257453 at 2.73 a resolution
|
| 60 | d1q45a_ |
|
not modelled |
24.2 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 61 | c3ohmB_ |
|
not modelled |
22.4 |
18 |
PDB header:signaling protein / hydrolase
Chain: B: PDB Molecule:1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase
PDBTitle: crystal structure of activated g alpha q bound to its effector2 phospholipase c beta 3
|
| 62 | c1djyB_ |
|
not modelled |
22.0 |
28 |
PDB header:lipid degradation
Chain: B: PDB Molecule:phosphoinositide-specific phospholipase c,
PDBTitle: phosphoinositide-specific phospholipase c-delta1 from rat2 complexed with inositol-2,4,5-trisphosphate
|
| 63 | c3bbnD_ |
|
not modelled |
22.0 |
27 |
PDB header:ribosome
Chain: D: PDB Molecule:ribosomal protein s4;
PDBTitle: homology model for the spinach chloroplast 30s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome.
|
| 64 | d1mn3a_ |
|
not modelled |
20.6 |
23 |
Fold:RuvA C-terminal domain-like
Superfamily:UBA-like
Family:CUE domain |
| 65 | c1bknA_ |
|
not modelled |
20.4 |
25 |
PDB header:dna repair
Chain: A: PDB Molecule:mutl;
PDBTitle: crystal structure of an n-terminal 40kd fragment of e. coli2 dna mismatch repair protein mutl
|
| 66 | d1geea_ |
|
not modelled |
19.9 |
30 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 67 | d1gtea2 |
|
not modelled |
19.8 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 68 | c1ddxA_ |
|
not modelled |
19.7 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein (prostaglandin h2 synthase-2);
PDBTitle: crystal structure of a mixture of arachidonic acid and prostaglandin2 bound to the cyclooxygenase active site of cox-2: prostaglandin3 structure
|
| 69 | c3pghD_ |
|
not modelled |
19.4 |
19 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:cyclooxygenase-2;
PDBTitle: cyclooxygenase-2 (prostaglandin synthase-2) complexed with a non-2 selective inhibitor, flurbiprofen
|
| 70 | c2fjuB_ |
|
not modelled |
18.9 |
18 |
PDB header:signaling protein,apoptosis/hydrolase
Chain: B: PDB Molecule:1-phosphatidylinositol-4,5-bisphosphate
PDBTitle: activated rac1 bound to its effector phospholipase c beta 2
|
| 71 | c3gdbA_ |
|
not modelled |
18.9 |
27 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative uncharacterized protein spr0440;
PDBTitle: crystal structure of spr0440 glycoside hydrolase domain,2 endo-d from streptococcus pneumoniae r6
|
| 72 | d1cvua1 |
|
not modelled |
18.3 |
19 |
Fold:Heme-dependent peroxidases
Superfamily:Heme-dependent peroxidases
Family:Myeloperoxidase-like |
| 73 | c3ivuB_ |
|
not modelled |
18.3 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:homocitrate synthase, mitochondrial;
PDBTitle: homocitrate synthase lys4 bound to 2-og
|
| 74 | d1vyua2 |
|
not modelled |
18.2 |
13 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 75 | c1pggB_ |
|
not modelled |
18.2 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:prostaglandin h2 synthase-1;
PDBTitle: prostaglandin h2 synthase-1 complexed with 1-(4-iodobenzoyl)-5-2 methoxy-2-methylindole-3-acetic acid (iodoindomethacin), trans model
|
| 76 | c1ht8B_ |
|
not modelled |
18.2 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:prostaglandin h2 synthase-1;
PDBTitle: the 2.7 angstrom resolution model of ovine cox-1 complexed with2 alclofenac
|
| 77 | c3kvoB_ |
|
not modelled |
18.1 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:hydroxysteroid dehydrogenase-like protein 2;
PDBTitle: crystal structure of the catalytic domain of human hydroxysteroid2 dehydrogenase like 2 (hsdl2)
|
| 78 | c1y2iC_ |
|
not modelled |
17.2 |
11 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:hypothetical protein s0862;
PDBTitle: crystal structure of mcsg target apc27401 from shigella2 flexneri
|
| 79 | d1y2ia_ |
|
not modelled |
17.2 |
11 |
Fold:Dodecin subunit-like
Superfamily:YbjQ-like
Family:YbjQ-like |
| 80 | c2w91A_ |
|
not modelled |
16.8 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:endo-beta-n-acetylglucosaminidase d;
PDBTitle: structure of a streptococcus pneumoniae family 85 glycoside2 hydrolase, endo-d.
|
| 81 | c2oyuP_ |
|
not modelled |
16.7 |
14 |
PDB header:oxidoreductase
Chain: P: PDB Molecule:prostaglandin g/h synthase 1;
PDBTitle: indomethacin-(s)-alpha-ethyl-ethanolamide bound to cyclooxygenase-1
|
| 82 | c2qr6A_ |
|
not modelled |
16.6 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:imp dehydrogenase/gmp reductase;
PDBTitle: crystal structure of imp dehydrogenase/gmp reductase-like protein2 (np_599840.1) from corynebacterium glutamicum atcc 13032 kitasato at3 1.50 a resolution
|
| 83 | c3qr0A_ |
|
not modelled |
16.5 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:phospholipase c-beta (plc-beta);
PDBTitle: crystal structure of s. officinalis plc21
|
| 84 | c1zfjA_ |
|
not modelled |
16.1 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine monophosphate dehydrogenase;
PDBTitle: inosine monophosphate dehydrogenase (impdh; ec 1.1.1.205) from2 streptococcus pyogenes
|
| 85 | d1o70a2 |
|
not modelled |
16.1 |
9 |
Fold:FAS1 domain
Superfamily:FAS1 domain
Family:FAS1 domain |
| 86 | c3mdoB_ |
|
not modelled |
15.7 |
15 |
PDB header:ligase
Chain: B: PDB Molecule:putative phosphoribosylformylglycinamidine cyclo-ligase;
PDBTitle: crystal structure of a putative phosphoribosylformylglycinamidine2 cyclo-ligase (bdi_2101) from parabacteroides distasonis atcc 8503 at3 1.91 a resolution
|
| 87 | c2a7rD_ |
|
not modelled |
15.7 |
22 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:gmp reductase 2;
PDBTitle: crystal structure of human guanosine monophosphate2 reductase 2 (gmpr2)
|
| 88 | c3gafF_ |
|
not modelled |
15.2 |
23 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:7-alpha-hydroxysteroid dehydrogenase;
PDBTitle: 2.2a crystal structure of 7-alpha-hydroxysteroid2 dehydrogenase from brucella melitensis
|
| 89 | d2cu0a1 |
|
not modelled |
14.9 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Inosine monophosphate dehydrogenase (IMPDH)
Family:Inosine monophosphate dehydrogenase (IMPDH) |
| 90 | c2kheA_ |
|
not modelled |
14.9 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:toxin-like protein;
PDBTitle: solution structure of the bacterial toxin rele from thermus2 thermophilus hb8
|
| 91 | d1w6ua_ |
|
not modelled |
14.8 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 92 | d1uzma1 |
|
not modelled |
14.6 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 93 | d1q4ga1 |
|
not modelled |
14.4 |
14 |
Fold:Heme-dependent peroxidases
Superfamily:Heme-dependent peroxidases
Family:Myeloperoxidase-like |
| 94 | d1pvna1 |
|
not modelled |
14.2 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Inosine monophosphate dehydrogenase (IMPDH)
Family:Inosine monophosphate dehydrogenase (IMPDH) |
| 95 | d2jn4a1 |
|
not modelled |
14.1 |
21 |
Fold:NifT/FixU barrel-like
Superfamily:NifT/FixU-like
Family:NifT/FixU |
| 96 | c2jn4A_ |
|
not modelled |
14.1 |
21 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein fixu, nift;
PDBTitle: solution nmr structure of protein rp4601 from2 rhodopseudomonas palustris. northeast structural genomics3 consortium target rpt2; ontario center for structural4 proteomics target rp4601.
|
| 97 | d1zfja1 |
|
not modelled |
14.0 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Inosine monophosphate dehydrogenase (IMPDH)
Family:Inosine monophosphate dehydrogenase (IMPDH) |
| 98 | c3rihB_ |
|
not modelled |
14.0 |
28 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:short chain dehydrogenase or reductase;
PDBTitle: crystal structure of a putative short chain dehydrogenase or reductase2 from mycobacterium abscessus
|
| 99 | c1d2vD_ |
|
not modelled |
14.0 |
19 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:myeloperoxidase;
PDBTitle: crystal structure of bromide-bound human myeloperoxidase isoform c at2 ph 5.5
|