1 c3efhB_
100.0
49
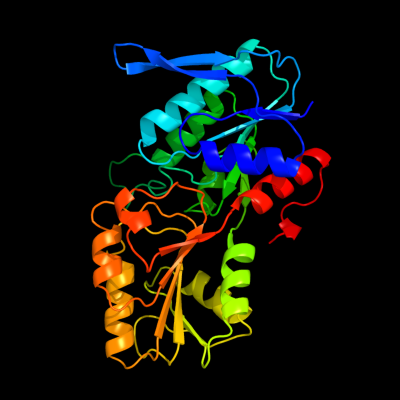
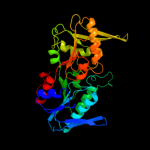
PDB header: transferaseChain: B: PDB Molecule: ribose-phosphate pyrophosphokinase 1;PDBTitle: crystal structure of human phosphoribosyl pyrophosphate2 synthetase 1
2 c1dkrB_
100.0
51
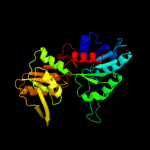
PDB header: transferaseChain: B: PDB Molecule: phosphoribosyl pyrophosphate synthetase;PDBTitle: crystal structures of bacillus subtilis phosphoribosylpyrophosphate2 synthetase: molecular basis of allosteric inhibition and activation.
3 c3dahB_
100.0
63
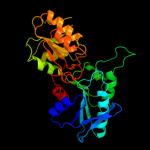
PDB header: transferaseChain: B: PDB Molecule: ribose-phosphate pyrophosphokinase;PDBTitle: 2.3 a crystal structure of ribose-phosphate pyrophosphokinase from2 burkholderia pseudomallei
4 c2c4kD_
100.0
41
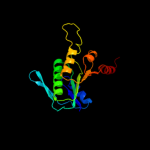
PDB header: regulatory proteinChain: D: PDB Molecule: phosphoribosyl pyrophosphate synthetase-PDBTitle: crystal structure of human phosphoribosylpyrophosphate2 synthetase-associated protein 39 (pap39)
5 c3lpnB_
100.0
31
PDB header: transferaseChain: B: PDB Molecule: ribose-phosphate pyrophosphokinase;PDBTitle: crystal structure of the phosphoribosylpyrophosphate (prpp) synthetase2 from thermoplasma volcanium in complex with an atp analog (ampcpp).
6 c1u9yD_
100.0
34
PDB header: transferaseChain: D: PDB Molecule: ribose-phosphate pyrophosphokinase;PDBTitle: crystal structure of phosphoribosyl diphosphate synthase2 from methanocaldococcus jannaschii
7 d1dkua1
100.0
55
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosylpyrophosphate synthetase-like8 d1u9ya1
100.0
31
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosylpyrophosphate synthetase-like9 d2c4ka1
100.0
42
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosylpyrophosphate synthetase-like10 d2c4ka2
100.0
39
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosylpyrophosphate synthetase-like11 d1dkua2
100.0
48
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosylpyrophosphate synthetase-like12 d1u9ya2
100.0
36
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosylpyrophosphate synthetase-like13 d1wd5a_
99.7
27
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)14 d1zn7a1
99.6
18
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)15 c3o7mD_
99.6
18
PDB header: transferaseChain: D: PDB Molecule: hypoxanthine phosphoribosyltransferase;PDBTitle: 1.98 angstrom resolution crystal structure of a hypoxanthine-guanine2 phosphoribosyltransferase (hpt-2) from bacillus anthracis str. 'ames3 ancestor'
16 c3dezA_
99.6
18
PDB header: transferaseChain: A: PDB Molecule: orotate phosphoribosyltransferase;PDBTitle: crystal structure of orotate phosphoribosyltransferase from2 streptococcus mutans
17 d1g9sa_
99.5
18
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)18 c2dy0A_
99.5
20
PDB header: transferaseChain: A: PDB Molecule: adenine phosphoribosyltransferase;PDBTitle: crystal structure of project jw0458 from escherichia coli
19 c2yzkC_
99.5
22
PDB header: transferaseChain: C: PDB Molecule: orotate phosphoribosyltransferase;PDBTitle: crystal structure of orotate phosphoribosyltransferase from2 aeropyrum pernix
20 c1yfzA_
99.5
17
PDB header: transferaseChain: A: PDB Molecule: hypoxanthine-guanine phosphoribosyltransferase;PDBTitle: novel imp binding in feedback inhibition of hypoxanthine-guanine2 phosphoribosyltransferase from thermoanaerobacter tengcongensis
21 d1yfza1
not modelled
99.5
17
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)22 d1vdma1
not modelled
99.5
27
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)23 d1z7ga1
not modelled
99.5
18
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)24 d1hgxa_
not modelled
99.5
20
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)25 d2aeea1
not modelled
99.5
17
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)26 d1ecfa1
not modelled
99.5
20
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)27 c3kb8A_
not modelled
99.5
22
PDB header: transferaseChain: A: PDB Molecule: hypoxanthine phosphoribosyltransferase;PDBTitle: 2.09 angstrom resolution structure of a hypoxanthine-guanine2 phosphoribosyltransferase (hpt-1) from bacillus anthracis str. 'ames3 ancestor' in complex with gmp
28 c2ywtA_
not modelled
99.5
18
PDB header: transferaseChain: A: PDB Molecule: hypoxanthine-guanine phosphoribosyltransferase;PDBTitle: crystal structure of hypoxanthine-guanine2 phosphoribosyltransferase with gmp from thermus3 thermophilus hb8
29 d1j7ja_
not modelled
99.4
20
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)30 d1l1qa_
not modelled
99.4
16
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)31 d1ufra_
not modelled
99.4
22
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)32 d1gph11
not modelled
99.4
19
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)33 d1cjba_
not modelled
99.4
22
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)34 d1g2qa_
not modelled
99.4
18
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)35 d1tc1a_
not modelled
99.4
13
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)36 d1p17b_
not modelled
99.4
17
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)37 d1y0ba1
not modelled
99.4
11
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)38 c2wnsB_
not modelled
99.4
17
PDB header: transferaseChain: B: PDB Molecule: orotate phosphoribosyltransferase;PDBTitle: human orotate phosphoribosyltransferase (oprtase) domain of2 uridine 5'-monophosphate synthase (umps) in complex with3 its substrate orotidine 5'-monophosphate (omp)
39 d1vcha1
not modelled
99.3
17
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)40 d1mzva_
not modelled
99.3
19
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)41 d1w30a_
not modelled
99.3
17
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)42 d1a3ca_
not modelled
99.3
21
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)43 d2igba1
not modelled
99.3
22
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)44 d1qb7a_
not modelled
99.3
19
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)45 c1ecjB_
not modelled
99.3
18
PDB header: transferaseChain: B: PDB Molecule: glutamine phosphoribosylpyrophosphatePDBTitle: escherichia coli glutamine phosphoribosylpyrophosphate2 (prpp) amidotransferase complexed with 2 amp per tetramer
46 c2p1zA_
not modelled
99.3
20
PDB header: transferaseChain: A: PDB Molecule: phosphoribosyltransferase;PDBTitle: crystal structure of phosphoribosyltransferase from corynebacterium2 diphtheriae
47 c1gph1_
not modelled
99.3
22
PDB header: transferase(glutamine amidotransferase)Chain: 1: PDB Molecule: glutamine phosphoribosyl-pyrophosphate amidotransferase;PDBTitle: structure of the allosteric regulatory enzyme of purine biosynthesis
48 d1pzma_
not modelled
99.2
13
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)49 d1o57a2
not modelled
99.2
19
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)50 c1pzmB_
not modelled
99.2
13
PDB header: transferaseChain: B: PDB Molecule: hypoxanthine-guanine phosphoribosyltransferase;PDBTitle: crystal structure of hgprt-ase from leishmania tarentolae in complex2 with gmp
51 c3m3hA_
not modelled
99.2
19
PDB header: transferaseChain: A: PDB Molecule: orotate phosphoribosyltransferase;PDBTitle: 1.75 angstrom resolution crystal structure of an orotate2 phosphoribosyltransferase from bacillus anthracis str. 'ames3 ancestor'
52 d1fsga_
not modelled
99.2
14
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)53 d1lh0a_
not modelled
99.2
18
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)54 c1o57A_
not modelled
99.1
20
PDB header: dna binding proteinChain: A: PDB Molecule: pur operon repressor;PDBTitle: crystal structure of the purine operon repressor of2 bacillus subtilis
55 c2jbhA_
not modelled
99.1
17
PDB header: transferaseChain: A: PDB Molecule: phosphoribosyltransferase domain-containing protein 1;PDBTitle: human phosphoribosyl transferase domain containing 1
56 c3mjdA_
not modelled
99.0
17
PDB header: transferaseChain: A: PDB Molecule: orotate phosphoribosyltransferase;PDBTitle: 1.9 angstrom crystal structure of orotate2 phosphoribosyltransferase (pyre) francisella tularensis.
57 c2jkzB_
not modelled
99.0
26
PDB header: transferaseChain: B: PDB Molecule: hypoxanthine-guanine phosphoribosyltransferase;PDBTitle: saccharomyces cerevisiae hypoxanthine-guanine2 phosphoribosyltransferase in complex with gmp (guanosine 5'3 -monophosphate) (orthorhombic crystal form)
58 c2przB_
not modelled
99.0
16
PDB header: transferaseChain: B: PDB Molecule: orotate phosphoribosyltransferase 1;PDBTitle: s. cerevisiae orotate phosphoribosyltransferase complexed2 with omp
59 c3qw4B_
not modelled
98.9
20
PDB header: transferase, lyaseChain: B: PDB Molecule: ump synthase;PDBTitle: structure of leishmania donovani ump synthase
60 d1o5oa_
not modelled
98.9
26
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)61 d1i5ea_
not modelled
98.9
25
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)62 c2ehjA_
not modelled
98.9
16
PDB header: transferaseChain: A: PDB Molecule: uracil phosphoribosyltransferase;PDBTitle: structure of uracil phosphoribosyl transferase
63 c2e55D_
not modelled
98.9
17
PDB header: transferaseChain: D: PDB Molecule: uracil phosphoribosyltransferase;PDBTitle: structure of aq2163 protein from aquifex aeolicus
64 c3n2lA_
not modelled
98.8
17
PDB header: transferaseChain: A: PDB Molecule: orotate phosphoribosyltransferase;PDBTitle: 2.1 angstrom resolution crystal structure of an orotate2 phosphoribosyltransferase (pyre) from vibrio cholerae o1 biovar eltor3 str. n16961
65 d1nula_
not modelled
98.8
21
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)66 c3dmpD_
not modelled
98.7
17
PDB header: transferaseChain: D: PDB Molecule: uracil phosphoribosyltransferase;PDBTitle: 2.6 a crystal structure of uracil phosphoribosyltransferase2 from burkholderia pseudomallei
67 d1xtta1
not modelled
98.6
17
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)68 d1bd3a_
not modelled
98.6
18
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)69 d1v9sa1
not modelled
98.5
27
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)70 d1dqna_
not modelled
98.5
16
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)71 c1zuwA_
not modelled
88.3
16
PDB header: isomeraseChain: A: PDB Molecule: glutamate racemase 1;PDBTitle: crystal structure of b.subtilis glutamate racemase (race) with d-glu
72 c2zskA_
not modelled
86.7
13
PDB header: unknown functionChain: A: PDB Molecule: 226aa long hypothetical aspartate racemase;PDBTitle: crystal structure of ph1733, an aspartate racemase2 homologue, from pyrococcus horikoshii ot3
73 c1sy7B_
not modelled
85.1
17
PDB header: oxidoreductaseChain: B: PDB Molecule: catalase 1;PDBTitle: crystal structure of the catalase-1 from neurospora crassa, native2 structure at 1.75a resolution.
74 c2eggA_
not modelled
83.9
15
PDB header: oxidoreductaseChain: A: PDB Molecule: shikimate 5-dehydrogenase;PDBTitle: crystal structure of shikimate 5-dehydrogenase (aroe) from2 geobacillus kaustophilus
75 c2hk8B_
not modelled
82.5
17
PDB header: oxidoreductaseChain: B: PDB Molecule: shikimate dehydrogenase;PDBTitle: crystal structure of shikimate dehydrogenase from aquifex2 aeolicus at 2.35 angstrom resolution
76 c2xecD_
not modelled
82.0
19
PDB header: isomeraseChain: D: PDB Molecule: putative maleate isomerase;PDBTitle: nocardia farcinica maleate cis-trans isomerase bound to2 tris
77 c3tfxB_
not modelled
80.6
16
PDB header: lyaseChain: B: PDB Molecule: orotidine 5'-phosphate decarboxylase;PDBTitle: crystal structure of orotidine 5'-phosphate decarboxylase from2 lactobacillus acidophilus
78 c2dx7B_
not modelled
79.9
19
PDB header: isomeraseChain: B: PDB Molecule: aspartate racemase;PDBTitle: crystal structure of pyrococcus horikoshii ot3 aspartate racemase2 complex with citric acid
79 c2jfnA_
not modelled
78.2
10
PDB header: isomeraseChain: A: PDB Molecule: glutamate racemase;PDBTitle: crystal structure of escherichia coli glutamate racemase2 in complex with l-glutamate and activator udp-murnac-ala
80 c3fbtB_
not modelled
77.3
14
PDB header: oxidoreductase, lyaseChain: B: PDB Molecule: chorismate mutase and shikimate 5-dehydrogenasePDBTitle: crystal structure of a chorismate mutase/shikimate 5-2 dehydrogenase fusion protein from clostridium3 acetobutylicum
81 c3hfrA_
not modelled
77.2
14
PDB header: isomeraseChain: A: PDB Molecule: glutamate racemase;PDBTitle: crystal structure of glutamate racemase from listeria monocytogenes
82 d1gtea3
not modelled
77.0
20
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: C-terminal domain of adrenodoxin reductase-like83 c1vi2B_
not modelled
76.6
17
PDB header: oxidoreductaseChain: B: PDB Molecule: shikimate 5-dehydrogenase 2;PDBTitle: crystal structure of shikimate-5-dehydrogenase with nad
84 c2cnwF_
not modelled
75.6
18
PDB header: signal recognitionChain: F: PDB Molecule: cell division protein ftsy;PDBTitle: gdpalf4 complex of the srp gtpases ffh and ftsy
85 c3r2uC_
not modelled
72.7
26
PDB header: hydrolaseChain: C: PDB Molecule: metallo-beta-lactamase family protein;PDBTitle: 2.1 angstrom resolution crystal structure of metallo-beta-lactamase2 from staphylococcus aureus subsp. aureus col
86 c2px0D_
not modelled
72.4
11
PDB header: biosynthetic proteinChain: D: PDB Molecule: flagellar biosynthesis protein flhf;PDBTitle: crystal structure of flhf complexed with gmppnp/mg(2+)
87 c3donA_
not modelled
71.2
13
PDB header: oxidoreductaseChain: A: PDB Molecule: shikimate dehydrogenase;PDBTitle: crystal structure of shikimate dehydrogenase from staphylococcus2 epidermidis
88 c3tozA_
not modelled
71.1
15
PDB header: oxidoreductaseChain: A: PDB Molecule: shikimate dehydrogenase;PDBTitle: 2.2 angstrom crystal structure of shikimate 5-dehydrogenase from2 listeria monocytogenes in complex with nad.
89 d1vi2a1
not modelled
70.3
24
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Aminoacid dehydrogenase-like, C-terminal domain90 d1vmka_
not modelled
68.5
12
Fold: Phosphorylase/hydrolase-likeSuperfamily: Purine and uridine phosphorylasesFamily: Purine and uridine phosphorylases91 c3sg0A_
not modelled
68.4
12
PDB header: signaling proteinChain: A: PDB Molecule: extracellular ligand-binding receptor;PDBTitle: the crystal structure of an extracellular ligand-binding receptor from2 rhodopseudomonas palustris haa2
92 d1efpb_
not modelled
67.0
15
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: ETFP subunits93 d1losc_
not modelled
66.4
13
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Decarboxylase94 c3dmdA_
not modelled
66.4
13
PDB header: transport proteinChain: A: PDB Molecule: signal recognition particle receptor;PDBTitle: structures and conformations in solution of the signal recognition2 particle receptor from the archaeon pyrococcus furiosus
95 c3o8qB_
not modelled
65.9
18
PDB header: oxidoreductaseChain: B: PDB Molecule: shikimate 5-dehydrogenase i alpha;PDBTitle: 1.45 angstrom resolution crystal structure of shikimate 5-2 dehydrogenase (aroe) from vibrio cholerae
96 c3hp7A_
not modelled
65.0
31
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hemolysin, putative;PDBTitle: putative hemolysin from streptococcus thermophilus.
97 c2gzmB_
not modelled
64.9
16
PDB header: isomeraseChain: B: PDB Molecule: glutamate racemase;PDBTitle: crystal structure of the glutamate racemase from bacillus2 anthracis
98 d1vp8a_
not modelled
63.3
25
Fold: Pyruvate kinase C-terminal domain-likeSuperfamily: PK C-terminal domain-likeFamily: MTH1675-like99 c2dgdD_
not modelled
63.2
8
PDB header: lyaseChain: D: PDB Molecule: 223aa long hypothetical arylmalonate decarboxylase;PDBTitle: crystal structure of st0656, a function unknown protein from2 sulfolobus tokodaii
100 c3uagA_
not modelled
63.2
13
PDB header: ligaseChain: A: PDB Molecule: protein (udp-n-acetylmuramoyl-l-alanine:d-PDBTitle: udp-n-acetylmuramoyl-l-alanine:d-glutamate ligase
101 d1p77a1
not modelled
63.1
9
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Aminoacid dehydrogenase-like, C-terminal domain102 c3dm5A_
not modelled
62.1
13
PDB header: rna binding protein, transport proteinChain: A: PDB Molecule: signal recognition 54 kda protein;PDBTitle: structures of srp54 and srp19, the two proteins assembling2 the ribonucleic core of the signal recognition particle3 from the archaeon pyrococcus furiosus.
103 c3euaD_
not modelled
61.7
14
PDB header: isomeraseChain: D: PDB Molecule: putative fructose-aminoacid-6-phosphate deglycase;PDBTitle: crystal structure of a putative phosphosugar isomerase (bsu32610) from2 bacillus subtilis at 1.90 a resolution
104 c2a3nA_
not modelled
60.6
15
PDB header: sugar binding proteinChain: A: PDB Molecule: putative glucosamine-fructose-6-phosphate aminotransferase;PDBTitle: crystal structure of a putative glucosamine-fructose-6-phosphate2 aminotransferase (stm4540.s) from salmonella typhimurium lt2 at 1.353 a resolution
105 c3dfzB_
not modelled
60.3
15
PDB header: oxidoreductaseChain: B: PDB Molecule: precorrin-2 dehydrogenase;PDBTitle: sirc, precorrin-2 dehydrogenase
106 d1j8yf2
not modelled
60.1
10
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like107 d1eixa_
not modelled
59.8
17
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Decarboxylase108 c2xf4A_
not modelled
59.6
32
PDB header: hydrolaseChain: A: PDB Molecule: hydroxyacylglutathione hydrolase;PDBTitle: crystal structure of salmonella enterica serovar2 typhimurium ycbl
109 c3navB_
not modelled
59.6
15
PDB header: lyaseChain: B: PDB Molecule: tryptophan synthase alpha chain;PDBTitle: crystal structure of an alpha subunit of tryptophan synthase from2 vibrio cholerae o1 biovar el tor str. n16961
110 d1cjca1
not modelled
59.2
32
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: C-terminal domain of adrenodoxin reductase-like111 c3lubE_
not modelled
58.4
21
PDB header: hydrolaseChain: E: PDB Molecule: putative creatinine amidohydrolase;PDBTitle: crystal structure of putative creatinine amidohydrolase2 (yp_211512.1) from bacteroides fragilis nctc 9343 at 2.11 a3 resolution
112 c3la8A_
not modelled
58.2
16
PDB header: transferaseChain: A: PDB Molecule: putative purine nucleoside phosphorylase;PDBTitle: the crystal structure of smu.1229 from streptococcus mutans ua159
113 d1km4a_
not modelled
57.6
13
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Decarboxylase114 c3pwzA_
not modelled
57.3
17
PDB header: oxidoreductaseChain: A: PDB Molecule: shikimate dehydrogenase 3;PDBTitle: crystal structure of an ael1 enzyme from pseudomonas putida
115 c3ek2D_
not modelled
57.0
24
PDB header: oxidoreductaseChain: D: PDB Molecule: enoyl-(acyl-carrier-protein) reductase (nadh);PDBTitle: crystal structure of eonyl-(acyl carrier protein) reductase2 from burkholderia pseudomallei 1719b
116 c2j7pA_
not modelled
56.9
11
PDB header: signal recognitionChain: A: PDB Molecule: signal recognition particle protein;PDBTitle: gmppnp-stabilized ng domain complex of the srp gtpases ffh2 and ftsy
117 c3shoA_
not modelled
56.2
16
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator, rpir family;PDBTitle: crystal structure of rpir transcription factor from sphaerobacter2 thermophilus (sugar isomerase domain)
118 c3b9qA_
not modelled
55.5
17
PDB header: protein transportChain: A: PDB Molecule: chloroplast srp receptor homolog, alpha subunitPDBTitle: the crystal structure of cpftsy from arabidopsis thaliana
119 d1qsga_
not modelled
55.4
24
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases120 c3qw3B_
not modelled
55.4
16
PDB header: transferase, lyaseChain: B: PDB Molecule: orotidine-5-phosphate decarboxylase/orotatePDBTitle: structure of leishmania donovani omp decarboxylase