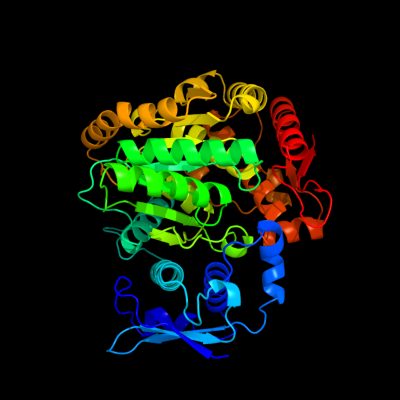
1 c3bq9A_
100.0
62
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: predicted rossmann fold nucleotide-binding domain-PDBTitle: crystal structure of predicted nucleotide-binding protein from2 idiomarina baltica os145
2 c3gh1A_
100.0
68
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: predicted nucleotide-binding protein;PDBTitle: crystal structure of predicted nucleotide-binding protein from vibrio2 cholerae
3 d1weka_
100.0
25
Fold: MCP/YpsA-likeSuperfamily: MCP/YpsA-likeFamily: MoCo carrier protein-like4 d1t35a_
100.0
17
Fold: MCP/YpsA-likeSuperfamily: MCP/YpsA-likeFamily: MoCo carrier protein-like5 c2q4dB_
100.0
19
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: lysine decarboxylase-like protein at5g11950;PDBTitle: ensemble refinement of the crystal structure of a lysine2 decarboxylase-like protein from arabidopsis thaliana gene at5g11950
6 d1ydha_
100.0
19
Fold: MCP/YpsA-likeSuperfamily: MCP/YpsA-likeFamily: MoCo carrier protein-like7 c3quaA_
100.0
19
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of a putative uncharacterized protein and possible2 molybdenum cofactor protein from mycobacterium smegmatis
8 c3sbxC_
100.0
20
PDB header: unknown functionChain: C: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of a putative uncharacterized protein from2 mycobacterium marinum bound to adenosine 5'-monophosphate amp
9 d2q4oa1
100.0
22
Fold: MCP/YpsA-likeSuperfamily: MCP/YpsA-likeFamily: MoCo carrier protein-like10 c2q4oA_
100.0
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein at2g37210/t2n18.3;PDBTitle: ensemble refinement of the crystal structure of a lysine2 decarboxylase-like protein from arabidopsis thaliana gene at2g37210
11 d1weha_
100.0
21
Fold: MCP/YpsA-likeSuperfamily: MCP/YpsA-likeFamily: MoCo carrier protein-like12 c1rcuB_
99.9
20
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: conserved hypothetical protein vt76;PDBTitle: x-ray structure of tm1055 northeast structural genomics2 consortium target vt76
13 d1rcua_
99.9
20
Fold: MCP/YpsA-likeSuperfamily: MCP/YpsA-likeFamily: MoCo carrier protein-like14 c2iz6A_
99.9
22
PDB header: metal transportChain: A: PDB Molecule: molybdenum cofactor carrier protein;PDBTitle: structure of the chlamydomonas rheinhardtii moco carrier2 protein
15 c3majA_
96.4
18
PDB header: dna binding proteinChain: A: PDB Molecule: dna processing chain a;PDBTitle: crystal structure of putative dna processing protein dpra from2 rhodopseudomonas palustris cga009
16 d1o1xa_
82.8
23
Fold: Ribose/Galactose isomerase RpiB/AlsBSuperfamily: Ribose/Galactose isomerase RpiB/AlsBFamily: Ribose/Galactose isomerase RpiB/AlsB17 d1e19a_
80.8
29
Fold: Carbamate kinase-likeSuperfamily: Carbamate kinase-likeFamily: Carbamate kinase18 c3c5yD_
77.3
18
PDB header: isomeraseChain: D: PDB Molecule: ribose/galactose isomerase;PDBTitle: crystal structure of a putative ribose 5-phosphate isomerase2 (saro_3514) from novosphingobium aromaticivorans dsm at 1.81 a3 resolution
19 c2ppwA_
75.3
18
PDB header: isomeraseChain: A: PDB Molecule: conserved domain protein;PDBTitle: the crystal structure of uncharacterized ribose 5-phosphate isomerase2 rpib from streptococcus pneumoniae
20 c3kzfC_
74.4
20
PDB header: transferaseChain: C: PDB Molecule: carbamate kinase;PDBTitle: structure of giardia carbamate kinase
21 c2fzvC_
not modelled
71.4
15
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: putative arsenical resistance protein;PDBTitle: crystal structure of an apo form of a flavin-binding protein from2 shigella flexneri
22 c3l86A_
not modelled
69.3
19
PDB header: transferaseChain: A: PDB Molecule: acetylglutamate kinase;PDBTitle: the crystal structure of smu.665 from streptococcus mutans ua159
23 c3he8A_
not modelled
67.8
20
PDB header: isomeraseChain: A: PDB Molecule: ribose-5-phosphate isomerase;PDBTitle: structural study of clostridium thermocellum ribose-5-phosphate2 isomerase b
24 c2e9yA_
not modelled
60.7
24
PDB header: transferaseChain: A: PDB Molecule: carbamate kinase;PDBTitle: crystal structure of project ape1968 from aeropyrum pernix k1
25 d1b7ba_
not modelled
59.0
27
Fold: Carbamate kinase-likeSuperfamily: Carbamate kinase-likeFamily: Carbamate kinase26 d2btya1
not modelled
58.9
19
Fold: Carbamate kinase-likeSuperfamily: Carbamate kinase-likeFamily: N-acetyl-l-glutamate kinase27 d2bufa1
not modelled
58.1
23
Fold: Carbamate kinase-likeSuperfamily: Carbamate kinase-likeFamily: N-acetyl-l-glutamate kinase28 c2rd5A_
not modelled
55.3
18
PDB header: protein bindingChain: A: PDB Molecule: acetylglutamate kinase-like protein;PDBTitle: structural basis for the regulation of n-acetylglutamate kinase by pii2 in arabidopsis thaliana
29 d2ij9a1
not modelled
53.9
18
Fold: Carbamate kinase-likeSuperfamily: Carbamate kinase-likeFamily: PyrH-like30 c2ogxB_
not modelled
53.7
28
PDB header: metal binding proteinChain: B: PDB Molecule: molybdenum storage protein subunit beta;PDBTitle: the crystal structure of the molybdenum storage protein from2 azotobacter vinelandii loaded with polyoxotungstates (wsto)
31 d2nu7b1
not modelled
50.8
7
Fold: Flavodoxin-likeSuperfamily: Succinyl-CoA synthetase domainsFamily: Succinyl-CoA synthetase domains32 c2w21A_
not modelled
50.7
16
PDB header: transferaseChain: A: PDB Molecule: glutamate 5-kinase;PDBTitle: crystal structure of the aminoacid kinase domain of the2 glutamate 5 kinase of escherichia coli.
33 c3nwyB_
not modelled
49.6
15
PDB header: transferaseChain: B: PDB Molecule: uridylate kinase;PDBTitle: structure and allosteric regulation of the uridine monophosphate2 kinase from mycobacterium tuberculosis
34 c2ekcA_
not modelled
49.5
24
PDB header: lyaseChain: A: PDB Molecule: tryptophan synthase alpha chain;PDBTitle: structural study of project id aq_1548 from aquifex aeolicus vf5
35 c3ek5A_
not modelled
49.0
10
PDB header: transferaseChain: A: PDB Molecule: uridylate kinase;PDBTitle: unique gtp-binding pocket and allostery of ump kinase from a gram-2 negative phytopathogen bacterium
36 d2akoa1
not modelled
48.5
27
Fold: Carbamate kinase-likeSuperfamily: Carbamate kinase-likeFamily: PyrH-like37 d1vhqa_
not modelled
45.1
24
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: DJ-1/PfpI38 c2l5aA_
not modelled
45.1
22
PDB header: nuclear proteinChain: A: PDB Molecule: histone h3-like centromeric protein cse4, protein scm3,PDBTitle: structural basis for recognition of centromere specific histone h32 variant by nonhistone scm3
39 c2jjxC_
not modelled
44.4
18
PDB header: transferaseChain: C: PDB Molecule: uridylate kinase;PDBTitle: the crystal structure of ump kinase from bacillus anthracis2 (ba1797)
40 d2ocda1
not modelled
44.0
13
Fold: Glutaminase/AsparaginaseSuperfamily: Glutaminase/AsparaginaseFamily: Glutaminase/Asparaginase41 d1z9da1
not modelled
43.0
10
Fold: Carbamate kinase-likeSuperfamily: Carbamate kinase-likeFamily: PyrH-like42 d1ikta_
not modelled
41.9
9
Fold: SCP-likeSuperfamily: SCP-likeFamily: Sterol carrier protein, SCP43 c3bn8A_
not modelled
41.3
27
PDB header: transport proteinChain: A: PDB Molecule: putative sterol carrier protein 2;PDBTitle: crystal structure of a putative sterol carrier protein type 2 (af1534)2 from archaeoglobus fulgidus dsm 4304 at 2.11 a resolution
44 c3o8nA_
not modelled
40.3
10
PDB header: transferaseChain: A: PDB Molecule: 6-phosphofructokinase, muscle type;PDBTitle: structure of phosphofructokinase from rabbit skeletal muscle
45 c3qd5B_
not modelled
39.8
15
PDB header: isomeraseChain: B: PDB Molecule: putative ribose-5-phosphate isomerase;PDBTitle: crystal structure of a putative ribose-5-phosphate isomerase from2 coccidioides immitis solved by combined iodide ion sad and mr
46 c2egxA_
not modelled
39.7
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative acetylglutamate kinase;PDBTitle: crystal structure of the putative acetylglutamate kinase from thermus2 thermophilus
47 d2fzva1
not modelled
38.9
14
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: NADPH-dependent FMN reductase48 c2j5gL_
not modelled
38.6
15
PDB header: hydrolaseChain: L: PDB Molecule: alr4455 protein;PDBTitle: the native structure of a beta-diketone hydrolase from the2 cyanobacterium anabaena sp. pcc 7120
49 c3onoA_
not modelled
37.5
18
PDB header: isomeraseChain: A: PDB Molecule: ribose/galactose isomerase;PDBTitle: crystal structure of ribose-5-phosphate isomerase lacab_rpib from2 vibrio parahaemolyticus
50 c2jzcA_
not modelled
36.0
8
PDB header: transferaseChain: A: PDB Molecule: udp-n-acetylglucosamine transferase subunitPDBTitle: nmr solution structure of alg13: the sugar donor subunit of2 a yeast n-acetylglucosamine transferase. northeast3 structural genomics consortium target yg1
51 c2j5tF_
not modelled
35.5
17
PDB header: transferaseChain: F: PDB Molecule: glutamate 5-kinase;PDBTitle: glutamate 5-kinase from escherichia coli complexed with2 glutamate
52 d1oaoa_
not modelled
35.3
11
Fold: Prismane protein-likeSuperfamily: Prismane protein-likeFamily: Carbon monoxide dehydrogenase53 d2brxa1
not modelled
34.2
18
Fold: Carbamate kinase-likeSuperfamily: Carbamate kinase-likeFamily: PyrH-like54 c3l3bA_
not modelled
33.6
24
PDB header: biosynthetic proteinChain: A: PDB Molecule: es1 family protein;PDBTitle: crystal structure of isoprenoid biosynthesis protein with2 amidotransferase-like domain from ehrlichia chaffeensis at 1.90a3 resolution
55 d1qopa_
not modelled
33.4
17
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes56 c3lo3E_
not modelled
32.9
13
PDB header: structure genomics, unknown functionChain: E: PDB Molecule: uncharacterized conserved protein;PDBTitle: the crystal structure of a conserved functionally unknown2 protein from colwellia psychrerythraea 34h.
57 c3m1pA_
not modelled
32.5
27
PDB header: isomeraseChain: A: PDB Molecule: ribose 5-phosphate isomerase;PDBTitle: structure of ribose 5-phosphate isomerase type b from trypanosoma2 cruzi, soaked with allose-6-phosphate
58 c3k7pA_
not modelled
32.5
27
PDB header: isomeraseChain: A: PDB Molecule: ribose 5-phosphate isomerase;PDBTitle: structure of mutant of ribose 5-phosphate isomerase type b from2 trypanosoma cruzi.
59 c2jbwB_
not modelled
32.0
29
PDB header: hydrolaseChain: B: PDB Molecule: 2,6-dihydroxy-pseudo-oxynicotine hydrolase;PDBTitle: crystal structure of the 2,6-dihydroxy-pseudo-oxynicotine2 hydrolase.
60 c2hihB_
not modelled
31.3
16
PDB header: hydrolaseChain: B: PDB Molecule: lipase 46 kda form;PDBTitle: crystal structure of staphylococcus hyicus lipase
61 c3l76B_
not modelled
30.8
20
PDB header: transferaseChain: B: PDB Molecule: aspartokinase;PDBTitle: crystal structure of aspartate kinase from synechocystis
62 c2va1A_
not modelled
28.6
18
PDB header: transferaseChain: A: PDB Molecule: uridylate kinase;PDBTitle: crystal structure of ump kinase from ureaplasma parvum
63 d1nn4a_
not modelled
28.3
26
Fold: Ribose/Galactose isomerase RpiB/AlsBSuperfamily: Ribose/Galactose isomerase RpiB/AlsBFamily: Ribose/Galactose isomerase RpiB/AlsB64 c2ogxA_
not modelled
28.0
30
PDB header: metal binding proteinChain: A: PDB Molecule: molybdenum storage protein subunit alpha;PDBTitle: the crystal structure of the molybdenum storage protein from2 azotobacter vinelandii loaded with polyoxotungstates (wsto)
65 d2jbwa1
not modelled
28.0
27
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: 2,6-dihydropseudooxynicotine hydrolase-like66 c3ab4K_
not modelled
27.5
12
PDB header: transferaseChain: K: PDB Molecule: aspartokinase;PDBTitle: crystal structure of feedback inhibition resistant mutant of aspartate2 kinase from corynebacterium glutamicum in complex with lysine and3 threonine
67 c3pm6B_
not modelled
27.4
14
PDB header: lyaseChain: B: PDB Molecule: putative fructose-bisphosphate aldolase;PDBTitle: crystal structure of a putative fructose-1,6-biphosphate aldolase from2 coccidioides immitis solved by combined sad mr
68 c2j4kC_
not modelled
27.3
21
PDB header: transferaseChain: C: PDB Molecule: uridylate kinase;PDBTitle: crystal structure of uridylate kinase from sulfolobus2 solfataricus in complex with ump to 2.2 angstrom3 resolution
69 d1ojta1
not modelled
27.3
38
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains70 d1qvwa_
not modelled
27.2
18
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: DJ-1/PfpI71 d1q1ra2
not modelled
25.1
19
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains72 c1ibtC_
not modelled
24.8
38
PDB header: lyaseChain: C: PDB Molecule: histidine decarboxylase beta chain;PDBTitle: structure of the d53,54n mutant of histidine decarboxylase at-170 c
73 c2l8kA_
not modelled
24.7
23
PDB header: viral proteinChain: A: PDB Molecule: non-structural protein 7;PDBTitle: nmr structure of the arterivirus nonstructural protein 7 alpha (nsp72 alpha)
74 d2fiua1
not modelled
24.6
26
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: Atu0297-like75 d1djqa3
not modelled
23.5
38
Fold: Nucleotide-binding domainSuperfamily: Nucleotide-binding domainFamily: N-terminal domain of adrenodoxin reductase-like76 d1lvla1
not modelled
23.5
38
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains77 c3nquA_
not modelled
23.2
34
PDB header: dna binding proteinChain: A: PDB Molecule: histone h3-like centromeric protein a;PDBTitle: crystal structure of partially trypsinized (cenp-a/h4)2 heterotetramer
78 d1hbnb1
not modelled
23.1
16
Fold: Methyl-coenzyme M reductase alpha and beta chain C-terminal domainSuperfamily: Methyl-coenzyme M reductase alpha and beta chain C-terminal domainFamily: Methyl-coenzyme M reductase alpha and beta chain C-terminal domain79 d4pgaa_
not modelled
22.6
29
Fold: Glutaminase/AsparaginaseSuperfamily: Glutaminase/AsparaginaseFamily: Glutaminase/Asparaginase80 d2pq6a1
not modelled
22.6
12
Fold: UDP-Glycosyltransferase/glycogen phosphorylaseSuperfamily: UDP-Glycosyltransferase/glycogen phosphorylaseFamily: UDPGT-like81 c3ll9A_
not modelled
22.5
20
PDB header: transferaseChain: A: PDB Molecule: isopentenyl phosphate kinase;PDBTitle: x-ray structures of isopentenyl phosphate kinase
82 d3lada1
not modelled
22.3
46
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains83 d1mqsa_
not modelled
22.0
21
Fold: Sec1/munc18-like (SM) proteinsSuperfamily: Sec1/munc18-like (SM) proteinsFamily: Sec1/munc18-like (SM) proteins84 c3opyB_
not modelled
22.0
21
PDB header: transferaseChain: B: PDB Molecule: 6-phosphofructo-1-kinase beta-subunit;PDBTitle: crystal structure of pichia pastoris phosphofructokinase in the t-2 state
85 c3opyH_
not modelled
22.0
21
PDB header: transferaseChain: H: PDB Molecule: 6-phosphofructo-1-kinase beta-subunit;PDBTitle: crystal structure of pichia pastoris phosphofructokinase in the t-2 state
86 c3navB_
not modelled
21.8
16
PDB header: lyaseChain: B: PDB Molecule: tryptophan synthase alpha chain;PDBTitle: crystal structure of an alpha subunit of tryptophan synthase from2 vibrio cholerae o1 biovar el tor str. n16961
87 d1id3a_
not modelled
21.4
30
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones88 d2j0wa1
not modelled
21.1
16
Fold: Carbamate kinase-likeSuperfamily: Carbamate kinase-likeFamily: PyrH-like89 c1jqkE_
not modelled
21.1
11
PDB header: oxidoreductaseChain: E: PDB Molecule: carbon monoxide dehydrogenase;PDBTitle: crystal structure of carbon monoxide dehydrogenase from2 rhodospirillum rubrum
90 d1jqka_
not modelled
21.1
11
Fold: Prismane protein-likeSuperfamily: Prismane protein-likeFamily: Carbon monoxide dehydrogenase91 d1onfa1
not modelled
21.0
46
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains92 d1acoa2
not modelled
20.5
14
Fold: Aconitase iron-sulfur domainSuperfamily: Aconitase iron-sulfur domainFamily: Aconitase iron-sulfur domain93 d1vdca1
not modelled
20.3
36
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains94 d2bona1
not modelled
20.3
26
Fold: NAD kinase/diacylglycerol kinase-likeSuperfamily: NAD kinase/diacylglycerol kinase-likeFamily: Diacylglycerol kinase-like95 d2bnea1
not modelled
20.2
10
Fold: Carbamate kinase-likeSuperfamily: Carbamate kinase-likeFamily: PyrH-like96 d2jgra1
not modelled
20.2
30
Fold: NAD kinase/diacylglycerol kinase-likeSuperfamily: NAD kinase/diacylglycerol kinase-likeFamily: Diacylglycerol kinase-like97 c1z6gA_
not modelled
20.2
36
PDB header: transferaseChain: A: PDB Molecule: guanylate kinase;PDBTitle: crystal structure of guanylate kinase from plasmodium falciparum
98 c2vvyC_
not modelled
19.9
19
PDB header: viral proteinChain: C: PDB Molecule: protein b15;PDBTitle: structure of vaccinia virus protein b14
99 d2vvpa1
not modelled
19.8
18
Fold: Ribose/Galactose isomerase RpiB/AlsBSuperfamily: Ribose/Galactose isomerase RpiB/AlsBFamily: Ribose/Galactose isomerase RpiB/AlsB