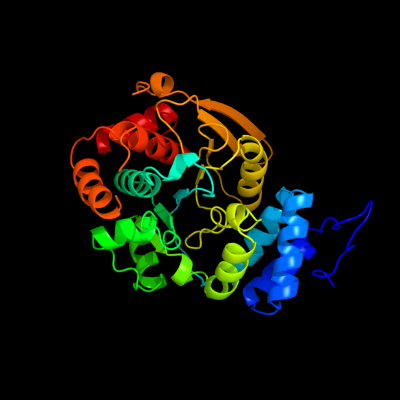
| 1 | c1v8gB_
|
|
 |
100.0 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:anthranilate phosphoribosyltransferase;
PDBTitle: crystal structure of anthranilate phosphoribosyltransferase2 (trpd) from thermus thermophilus hb8
|
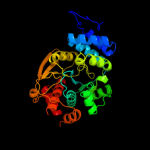
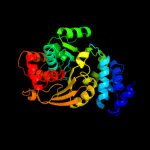
| 2 | c1o17A_
|
|
 |
100.0 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:anthranilate phosphoribosyltransferase;
PDBTitle: anthranilate phosphoribosyl-transferase (trpd)
|
| 3 | c1khdD_
|
|
 |
100.0 |
18 |
PDB header:transferase
Chain: D: PDB Molecule:anthranilate phosphoribosyltransferase;
PDBTitle: crystal structure analysis of the anthranilate2 phosphoribosyltransferase from erwinia carotovora at 1.93 resolution (current name, pectobacterium carotovorum)
|
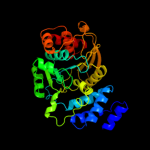
| 4 | c2bpqB_
|
|
 |
100.0 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:anthranilate phosphoribosyltransferase;
PDBTitle: anthranilate phosphoribosyltransferase (trpd) from2 mycobacterium tuberculosis (apo structure)
|
| 5 | c1vquB_
|
|
 |
100.0 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:anthranilate phosphoribosyltransferase 2;
PDBTitle: crystal structure of anthranilate phosphoribosyltransferase 22 (17130499) from nostoc sp. at 1.85 a resolution
|
| 6 | c3h5qA_
|
|
 |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:pyrimidine-nucleoside phosphorylase;
PDBTitle: crystal structure of a putative pyrimidine-nucleoside phosphorylase2 from staphylococcus aureus
|
| 7 | c1brwB_
|
|
 |
100.0 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:protein (pyrimidine nucleoside phosphorylase);
PDBTitle: the crystal structure of pyrimidine nucleoside2 phosphorylase in a closed conformation
|
| 8 | d1o17a2
|
|
 |
100.0 |
19 |
Fold:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Superfamily:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Family:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain |
| 9 | d2elca2
|
|
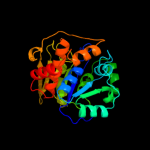 |
100.0 |
21 |
Fold:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Superfamily:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Family:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain |
| 10 | c2j0fC_
|
|
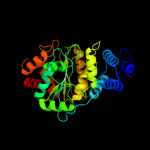 |
100.0 |
18 |
PDB header:transferase
Chain: C: PDB Molecule:thymidine phosphorylase;
PDBTitle: structural basis for non-competitive product inhibition in2 human thymidine phosphorylase: implication for drug design
|
| 11 | c2dsjA_
|
|
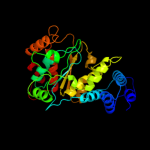 |
100.0 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:pyrimidine-nucleoside (thymidine) phosphorylase;
PDBTitle: crystal structure of project id tt0128 from thermus thermophilus hb8
|
| 12 | c1otpA_
|
|
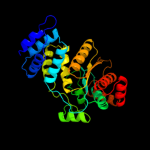 |
100.0 |
13 |
PDB header:phosphorylase
Chain: A: PDB Molecule:thymidine phosphorylase;
PDBTitle: structural and theoretical studies suggest domain movement produces an2 active conformation of thymidine phosphorylase
|
| 13 | d1khda2
|
|
 |
100.0 |
18 |
Fold:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Superfamily:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Family:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain |
| 14 | d1uoua2
|
|
 |
100.0 |
16 |
Fold:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Superfamily:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Family:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain |
| 15 | d1brwa2
|
|
 |
100.0 |
12 |
Fold:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Superfamily:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Family:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain |
| 16 | d2tpta2
|
|
 |
100.0 |
13 |
Fold:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Superfamily:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Family:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain |
| 17 | d1o17a1
|
|
 |
99.7 |
37 |
Fold:Methionine synthase domain-like
Superfamily:Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain
Family:Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain |
| 18 | d1khda1
|
|
 |
99.6 |
20 |
Fold:Methionine synthase domain-like
Superfamily:Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain
Family:Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain |
| 19 | d1brwa1
|
|
 |
99.6 |
21 |
Fold:Methionine synthase domain-like
Superfamily:Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain
Family:Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain |
| 20 | d2tpta1
|
|
 |
99.6 |
13 |
Fold:Methionine synthase domain-like
Superfamily:Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain
Family:Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain |
| 21 | d1v8ga1 |
|
not modelled |
99.5 |
25 |
Fold:Methionine synthase domain-like
Superfamily:Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain
Family:Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain |
| 22 | d1uoua1 |
|
not modelled |
99.5 |
23 |
Fold:Methionine synthase domain-like
Superfamily:Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain
Family:Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain |
| 23 | d2elca1 |
|
not modelled |
99.1 |
27 |
Fold:Methionine synthase domain-like
Superfamily:Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain
Family:Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain |
| 24 | d1vkma_ |
|
not modelled |
90.5 |
18 |
Fold:Indigoidine synthase A-like
Superfamily:Indigoidine synthase A-like
Family:Indigoidine synthase A-like |
| 25 | d1tuea_ |
|
not modelled |
72.9 |
16 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Extended AAA-ATPase domain |
| 26 | d1svma_ |
|
not modelled |
71.7 |
16 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Extended AAA-ATPase domain |
| 27 | d1kfia1 |
|
not modelled |
69.0 |
12 |
Fold:Phosphoglucomutase, first 3 domains
Superfamily:Phosphoglucomutase, first 3 domains
Family:Phosphoglucomutase, first 3 domains |
| 28 | c2kvcA_ |
|
not modelled |
68.6 |
13 |
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: solution structure of the mycobacterium tuberculosis protein rv0543c,2 a member of the duf3349 superfamily. seattle structural genomics3 center for infectious disease target mytud.17112.a
|
| 29 | d1p88a_ |
|
not modelled |
68.5 |
17 |
Fold:IF3-like
Superfamily:EPT/RTPC-like
Family:Enolpyruvate transferase, EPT |
| 30 | c2o0zA_ |
|
not modelled |
66.9 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:3-phosphoshikimate 1-carboxyvinyltransferase;
PDBTitle: mycobacterium tuberculosis epsp synthase in complex with2 product (eps)
|
| 31 | d2gc6a2 |
|
not modelled |
64.6 |
12 |
Fold:Ribokinase-like
Superfamily:MurD-like peptide ligases, catalytic domain
Family:Folylpolyglutamate synthetase |
| 32 | c2lkyA_ |
|
not modelled |
61.3 |
12 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution structure of msmeg_1053, the second duf3349 annotated protein2 in the genome of mycobacterium smegmatis, seattle structural genomics3 center for infectious disease target mysma.17112.b
|
| 33 | c2px0D_ |
|
not modelled |
60.6 |
14 |
PDB header:biosynthetic protein
Chain: D: PDB Molecule:flagellar biosynthesis protein flhf;
PDBTitle: crystal structure of flhf complexed with gmppnp/mg(2+)
|
| 34 | c3n2aA_ |
|
not modelled |
51.2 |
18 |
PDB header:ligase
Chain: A: PDB Molecule:bifunctional folylpolyglutamate synthase/dihydrofolate
PDBTitle: crystal structure of bifunctional folylpolyglutamate2 synthase/dihydrofolate synthase from yersinia pestis co92
|
| 35 | c2yvqA_ |
|
not modelled |
50.8 |
13 |
PDB header:ligase
Chain: A: PDB Molecule:carbamoyl-phosphate synthase;
PDBTitle: crystal structure of mgs domain of carbamoyl-phosphate2 synthetase from homo sapiens
|
| 36 | c3chgB_ |
|
not modelled |
47.8 |
21 |
PDB header:ligand binding protein
Chain: B: PDB Molecule:glycine betaine-binding protein;
PDBTitle: the compatible solute-binding protein opuac from bacillus2 subtilis in complex with dmsa
|
| 37 | c2gc6A_ |
|
not modelled |
47.2 |
18 |
PDB header:ligase
Chain: A: PDB Molecule:folylpolyglutamate synthase;
PDBTitle: s73a mutant of l. casei fpgs
|
| 38 | c1kfiA_ |
|
not modelled |
46.8 |
12 |
PDB header:isomerase
Chain: A: PDB Molecule:phosphoglucomutase 1;
PDBTitle: crystal structure of the exocytosis-sensitive2 phosphoprotein, pp63/parafusin (phosphoglucomutase) from3 paramecium
|
| 39 | d3pmga1 |
|
not modelled |
46.7 |
10 |
Fold:Phosphoglucomutase, first 3 domains
Superfamily:Phosphoglucomutase, first 3 domains
Family:Phosphoglucomutase, first 3 domains |
| 40 | d1j9ia_ |
|
not modelled |
45.6 |
11 |
Fold:Putative DNA-binding domain
Superfamily:Putative DNA-binding domain
Family:Terminase gpNU1 subunit domain |
| 41 | c2cnwF_ |
|
not modelled |
45.2 |
17 |
PDB header:signal recognition
Chain: F: PDB Molecule:cell division protein ftsy;
PDBTitle: gdpalf4 complex of the srp gtpases ffh and ftsy
|
| 42 | d1rf6a_ |
|
not modelled |
44.9 |
16 |
Fold:IF3-like
Superfamily:EPT/RTPC-like
Family:Enolpyruvate transferase, EPT |
| 43 | c1w96B_ |
|
not modelled |
43.1 |
8 |
PDB header:ligase
Chain: B: PDB Molecule:acetyl-coenzyme a carboxylase;
PDBTitle: crystal structure of biotin carboxylase domain of acetyl-2 coenzyme a carboxylase from saccharomyces cerevisiae in3 complex with soraphen a
|
| 44 | c3k2gA_ |
|
not modelled |
42.2 |
11 |
PDB header:resiniferatoxin binding protein
Chain: A: PDB Molecule:resiniferatoxin-binding, phosphotriesterase-
PDBTitle: crystal structure of a resiniferatoxin-binding protein from2 rhodobacter sphaeroides
|
| 45 | c1r71B_ |
|
not modelled |
41.5 |
13 |
PDB header:transcription/dna
Chain: B: PDB Molecule:transcriptional repressor protein korb;
PDBTitle: crystal structure of the dna binding domain of korb in2 complex with the operator dna
|
| 46 | d2jwda1 |
|
not modelled |
39.6 |
11 |
Fold:immunoglobulin/albumin-binding domain-like
Superfamily:Bacterial immunoglobulin/albumin-binding domains
Family:Immunoglobulin-binding protein A modules |
| 47 | c2pqdA_ |
|
not modelled |
39.5 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:3-phosphoshikimate 1-carboxyvinyltransferase;
PDBTitle: a100g cp4 epsps liganded with (r)-difluoromethyl tetrahedral reaction2 intermediate analog
|
| 48 | d1ukza_ |
|
not modelled |
38.9 |
14 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 49 | c3rmtB_ |
|
not modelled |
37.8 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:3-phosphoshikimate 1-carboxyvinyltransferase 1;
PDBTitle: crystal structure of putative 5-enolpyruvoylshikimate-3-phosphate2 synthase from bacillus halodurans c-125
|
| 50 | d1ejda_ |
|
not modelled |
37.5 |
11 |
Fold:IF3-like
Superfamily:EPT/RTPC-like
Family:Enolpyruvate transferase, EPT |
| 51 | d1tz9a_ |
|
not modelled |
36.9 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:UxuA-like |
| 52 | d1r71a_ |
|
not modelled |
36.7 |
14 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:KorB DNA-binding domain-like
Family:KorB DNA-binding domain-like |
| 53 | c2qguA_ |
|
not modelled |
36.2 |
2 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:probable signal peptide protein;
PDBTitle: three-dimensional structure of the phospholipid-binding protein from2 ralstonia solanacearum q8xv73_ralsq in complex with a phospholipid at3 the resolution 1.53 a. northeast structural genomics consortium4 target rsr89
|
| 54 | c3r38A_ |
|
not modelled |
35.8 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:udp-n-acetylglucosamine 1-carboxyvinyltransferase 1;
PDBTitle: 2.23 angstrom resolution crystal structure of udp-n-acetylglucosamine2 1-carboxyvinyltransferase (mura) from listeria monocytogenes egd-e
|
| 55 | c2oz5A_ |
|
not modelled |
35.0 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:phosphotyrosine protein phosphatase ptpb;
PDBTitle: crystal structure of mycobacterium tuberculosis protein tyrosine2 phosphatase ptpb in complex with the specific inhibitor omts
|
| 56 | c3jvpA_ |
|
not modelled |
34.7 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:ribulokinase;
PDBTitle: crystal structure of ribulokinase from bacillus halodurans
|
| 57 | c1w78A_ |
|
not modelled |
34.4 |
18 |
PDB header:synthase
Chain: A: PDB Molecule:folc bifunctional protein;
PDBTitle: e.coli folc in complex with dhpp and adp
|
| 58 | c3ewbX_ |
|
not modelled |
34.4 |
9 |
PDB header:transferase
Chain: X: PDB Molecule:2-isopropylmalate synthase;
PDBTitle: crystal structure of n-terminal domain of putative 2-2 isopropylmalate synthase from listeria monocytogenes
|
| 59 | c3eegB_ |
|
not modelled |
34.2 |
8 |
PDB header:transferase
Chain: B: PDB Molecule:2-isopropylmalate synthase;
PDBTitle: crystal structure of a 2-isopropylmalate synthase from2 cytophaga hutchinsonii
|
| 60 | c3bg3B_ |
|
not modelled |
34.1 |
10 |
PDB header:ligase
Chain: B: PDB Molecule:pyruvate carboxylase, mitochondrial;
PDBTitle: crystal structure of human pyruvate carboxylase (missing2 the biotin carboxylase domain at the n-terminus)
|
| 61 | c1nsfA_ |
|
not modelled |
33.9 |
17 |
PDB header:protein transport
Chain: A: PDB Molecule:n-ethylmaleimide sensitive factor;
PDBTitle: d2 hexamerization domain of n-ethylmaleimide sensitive factor (nsf)
|
| 62 | d1a0pa2 |
|
not modelled |
32.8 |
23 |
Fold:DNA breaking-rejoining enzymes
Superfamily:DNA breaking-rejoining enzymes
Family:Lambda integrase-like, catalytic core |
| 63 | d1uaea_ |
|
not modelled |
32.3 |
10 |
Fold:IF3-like
Superfamily:EPT/RTPC-like
Family:Enolpyruvate transferase, EPT |
| 64 | c1rr2A_ |
|
not modelled |
30.8 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:transcarboxylase 5s subunit;
PDBTitle: propionibacterium shermanii transcarboxylase 5s subunit bound to 2-2 ketobutyric acid
|
| 65 | c2yhsA_ |
|
not modelled |
30.7 |
18 |
PDB header:cell cycle
Chain: A: PDB Molecule:cell division protein ftsy;
PDBTitle: structure of the e. coli srp receptor ftsy
|
| 66 | d1g6sa_ |
|
not modelled |
30.6 |
18 |
Fold:IF3-like
Superfamily:EPT/RTPC-like
Family:Enolpyruvate transferase, EPT |
| 67 | d1ywfa1 |
|
not modelled |
29.9 |
15 |
Fold:(Phosphotyrosine protein) phosphatases II
Superfamily:(Phosphotyrosine protein) phosphatases II
Family:Mycobacterial PtpB-like |
| 68 | c2ip4A_ |
|
not modelled |
29.5 |
16 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylamine--glycine ligase;
PDBTitle: crystal structure of glycinamide ribonucleotide synthetase from2 thermus thermophilus hb8
|
| 69 | c3dnfB_ |
|
not modelled |
29.3 |
10 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:4-hydroxy-3-methylbut-2-enyl diphosphate reductase;
PDBTitle: structure of (e)-4-hydroxy-3-methyl-but-2-enyl diphosphate reductase,2 the terminal enzyme of the non-mevalonate pathway
|
| 70 | c2vosA_ |
|
not modelled |
28.6 |
14 |
PDB header:ligase
Chain: A: PDB Molecule:folylpolyglutamate synthase protein folc;
PDBTitle: mycobacterium tuberculosis folylpolyglutamate synthase2 complexed with adp
|
| 71 | c2iy3A_ |
|
not modelled |
27.6 |
15 |
PDB header:rna-binding
Chain: A: PDB Molecule:signal recognition particle protein ffh;
PDBTitle: structure of the e. coli signal recognition particle2 bound to a translating ribosome
|
| 72 | d1b8za_ |
|
not modelled |
26.9 |
10 |
Fold:IHF-like DNA-binding proteins
Superfamily:IHF-like DNA-binding proteins
Family:Prokaryotic DNA-bending protein |
| 73 | d1wb9a2 |
|
not modelled |
26.6 |
20 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:ABC transporter ATPase domain-like |
| 74 | c3ol4B_ |
|
not modelled |
26.5 |
14 |
PDB header:unknown function
Chain: B: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of a putative uncharacterized protein from2 mycobacterium smegmatis, an ortholog of rv0543c
|
| 75 | c2j37W_ |
|
not modelled |
26.4 |
13 |
PDB header:ribosome
Chain: W: PDB Molecule:signal recognition particle 54 kda protein
PDBTitle: model of mammalian srp bound to 80s rncs
|
| 76 | d1o5za2 |
|
not modelled |
25.9 |
21 |
Fold:Ribokinase-like
Superfamily:MurD-like peptide ligases, catalytic domain
Family:Folylpolyglutamate synthetase |
| 77 | c3bdkB_ |
|
not modelled |
25.6 |
10 |
PDB header:lyase
Chain: B: PDB Molecule:d-mannonate dehydratase;
PDBTitle: crystal structure of streptococcus suis mannonate2 dehydratase complexed with substrate analogue
|
| 78 | c3kc2A_ |
|
not modelled |
25.4 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:uncharacterized protein ykr070w;
PDBTitle: crystal structure of mitochondrial had-like phosphatase from2 saccharomyces cerevisiae
|
| 79 | c2dmxA_ |
|
not modelled |
25.0 |
8 |
PDB header:chaperone
Chain: A: PDB Molecule:dnaj homolog subfamily b member 8;
PDBTitle: solution structure of the j domain of dnaj homolog2 subfamily b member 8
|
| 80 | d2jdid1 |
|
not modelled |
24.9 |
12 |
Fold:Left-handed superhelix
Superfamily:C-terminal domain of alpha and beta subunits of F1 ATP synthase
Family:C-terminal domain of alpha and beta subunits of F1 ATP synthase |
| 81 | d1ak2a1 |
|
not modelled |
24.2 |
20 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 82 | d1ewqa2 |
|
not modelled |
24.1 |
12 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:ABC transporter ATPase domain-like |
| 83 | c2hjwA_ |
|
not modelled |
24.1 |
15 |
PDB header:ligase
Chain: A: PDB Molecule:acetyl-coa carboxylase 2;
PDBTitle: crystal structure of the bc domain of acc2
|
| 84 | d2afhe1 |
|
not modelled |
23.5 |
21 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
| 85 | d1fc2c_ |
|
not modelled |
23.5 |
14 |
Fold:immunoglobulin/albumin-binding domain-like
Superfamily:Bacterial immunoglobulin/albumin-binding domains
Family:Immunoglobulin-binding protein A modules |
| 86 | c3eihB_ |
|
not modelled |
23.3 |
14 |
PDB header:protein transport
Chain: B: PDB Molecule:vacuolar protein sorting-associated protein 4;
PDBTitle: crystal structure of s.cerevisiae vps4 in the presence of atpgammas
|
| 87 | c3ff5B_ |
|
not modelled |
23.0 |
15 |
PDB header:protein transport
Chain: B: PDB Molecule:peroxisomal biogenesis factor 14;
PDBTitle: crystal structure of the conserved n-terminal domain of the2 peroxisomal matrix-protein-import receptor, pex14p
|
| 88 | c1wbdA_ |
|
not modelled |
22.9 |
20 |
PDB header:dna-binding
Chain: A: PDB Molecule:dna mismatch repair protein muts;
PDBTitle: crystal structure of e. coli dna mismatch repair enzyme2 muts, e38q mutant, in complex with a g.t mismatch
|
| 89 | d1mula_ |
|
not modelled |
22.8 |
20 |
Fold:IHF-like DNA-binding proteins
Superfamily:IHF-like DNA-binding proteins
Family:Prokaryotic DNA-bending protein |
| 90 | d1sr9a2 |
|
not modelled |
22.6 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:HMGL-like |
| 91 | c2dn9A_ |
|
not modelled |
22.6 |
5 |
PDB header:apoptosis, chaperone
Chain: A: PDB Molecule:dnaj homolog subfamily a member 3;
PDBTitle: solution structure of j-domain from the dnaj homolog, human2 tid1 protein
|
| 92 | d1g5ta_ |
|
not modelled |
22.2 |
19 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:RecA protein-like (ATPase-domain) |
| 93 | c2w40C_ |
|
not modelled |
22.0 |
10 |
PDB header:transferase
Chain: C: PDB Molecule:glycerol kinase, putative;
PDBTitle: crystal structure of plasmodium falciparum glycerol kinase2 with bound glycerol
|
| 94 | c3roiA_ |
|
not modelled |
21.9 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:3-phosphoshikimate 1-carboxyvinyltransferase;
PDBTitle: 2.20 angstrom resolution structure of 3-phosphoshikimate 1-2 carboxyvinyltransferase (aroa) from coxiella burnetii
|
| 95 | c2iz1C_ |
|
not modelled |
21.9 |
15 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:6-phosphogluconate dehydrogenase, decarboxylating;
PDBTitle: 6pdh complexed with pex inhibitor synchrotron data
|
| 96 | d1tlha_ |
|
not modelled |
21.8 |
28 |
Fold:Anti-sigma factor AsiA
Superfamily:Anti-sigma factor AsiA
Family:Anti-sigma factor AsiA |
| 97 | c3endA_ |
|
not modelled |
21.7 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:light-independent protochlorophyllide reductase
PDBTitle: crystal structure of the l protein of rhodobacter2 sphaeroides light-independent protochlorophyllide3 reductase (bchl) with mgadp bound: a homologue of the4 nitrogenase fe protein
|
| 98 | d1rqba2 |
|
not modelled |
21.2 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:HMGL-like |
| 99 | d1g64b_ |
|
not modelled |
21.1 |
19 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:RecA protein-like (ATPase-domain) |
| 100 | d1iura_ |
|
not modelled |
21.1 |
9 |
Fold:Long alpha-hairpin
Superfamily:Chaperone J-domain
Family:Chaperone J-domain |
| 101 | c2yztA_ |
|
not modelled |
21.0 |
21 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative uncharacterized protein ttha1756;
PDBTitle: crystal structure of uncharacterized conserved protein from thermus2 thermophilus hb8
|
| 102 | c1djyB_ |
|
not modelled |
20.9 |
15 |
PDB header:lipid degradation
Chain: B: PDB Molecule:phosphoinositide-specific phospholipase c,
PDBTitle: phosphoinositide-specific phospholipase c-delta1 from rat2 complexed with inositol-2,4,5-trisphosphate
|
| 103 | d1gvnb_ |
|
not modelled |
20.5 |
11 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Plasmid maintenance system epsilon/zeta, toxin zeta subunit |
| 104 | c1o5zA_ |
|
not modelled |
20.3 |
21 |
PDB header:ligase
Chain: A: PDB Molecule:folylpolyglutamate synthase/dihydrofolate synthase;
PDBTitle: crystal structure of folylpolyglutamate synthase (tm0166) from2 thermotoga maritima at 2.10 a resolution
|