| 1 | c3d2rB_ |
|
 |
100.0 |
12 |
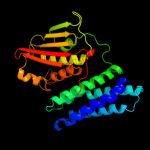
PDB header:transferase
Chain: B: PDB Molecule:[pyruvate dehydrogenase [lipoamide]] kinase isozyme 4;
PDBTitle: crystal structure of pyruvate dehydrogenase kinase isoform 4 in2 complex with adp
|
| 2 | c2q8fA_ |
|
 |
100.0 |
15 |
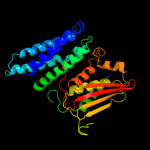
PDB header:transferase
Chain: A: PDB Molecule:[pyruvate dehydrogenase [lipoamide]] kinase isozyme 1;
PDBTitle: structure of pyruvate dehydrogenase kinase isoform 1
|
| 3 | c2bu8A_ |
|
 |
100.0 |
13 |
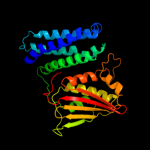
PDB header:transferase
Chain: A: PDB Molecule:pyruvate dehydrogenase kinase isoenzyme 2;
PDBTitle: crystal structures of human pyruvate dehydrogenase kinase 22 containing physiological and synthetic ligands
|
| 4 | c1y8oA_ |
|
 |
100.0 |
13 |
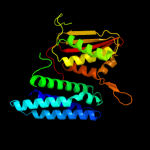
PDB header:transferase
Chain: A: PDB Molecule:[pyruvate dehydrogenase [lipoamide]] kinase isozyme 3;
PDBTitle: crystal structure of the pdk3-l2 complex
|
| 5 | c3d36B_ |
|
 |
100.0 |
20 |
PDB header:transferase/transferase inhibitor
Chain: B: PDB Molecule:sporulation kinase b;
PDBTitle: how to switch off a histidine kinase: crystal structure of2 geobacillus stearothermophilus kinb with the inhibitor sda
|
| 6 | c2c2aA_ |
|
 |
100.0 |
26 |
PDB header:transferase
Chain: A: PDB Molecule:sensor histidine kinase;
PDBTitle: structure of the entire cytoplasmic portion of a sensor2 histidine kinase protein
|
| 7 | c1gjvA_ |
|
 |
100.0 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:[3-methyl-2-oxobutanoate dehydrogenase
PDBTitle: branched-chain alpha-ketoacid dehydrogenase kinase (bck)2 complxed with atp-gamma-s
|
| 8 | d1bxda_ |
|
 |
100.0 |
100 |
Fold:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Superfamily:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Family:Histidine kinase |
| 9 | c3a0rA_ |
|
 |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:sensor protein;
PDBTitle: crystal structure of histidine kinase thka (tm1359) in complex with2 response regulator protein trra (tm1360)
|
| 10 | d1jm6a2 |
|
 |
100.0 |
20 |
Fold:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Superfamily:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Family:alpha-ketoacid dehydrogenase kinase, C-terminal domain |
| 11 | c1b3qA_ |
|
 |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:protein (chemotaxis protein chea);
PDBTitle: crystal structure of chea-289, a signal transducing histidine kinase
|
| 12 | c3a0tA_ |
|
 |
100.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:sensor protein;
PDBTitle: catalytic domain of histidine kinase thka (tm1359) in2 complex with adp and mg ion (trigonal)
|
| 13 | d1id0a_ |
|
 |
100.0 |
24 |
Fold:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Superfamily:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Family:Histidine kinase |
| 14 | d2c2aa2 |
|
 |
99.9 |
24 |
Fold:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Superfamily:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Family:Histidine kinase |
| 15 | d1gkza2 |
|
 |
99.9 |
16 |
Fold:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Superfamily:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Family:alpha-ketoacid dehydrogenase kinase, C-terminal domain |
| 16 | d1ysra1 |
|
 |
99.9 |
24 |
Fold:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Superfamily:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Family:Histidine kinase |
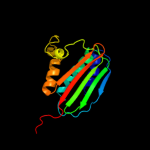
| 17 | c2ch4A_ |
|
 |
99.9 |
27 |
PDB header:transferase/chemotaxis
Chain: A: PDB Molecule:chemotaxis protein chea;
PDBTitle: complex between bacterial chemotaxis histidine kinase chea2 domains p4 and p5 and receptor-adaptor protein chew
|
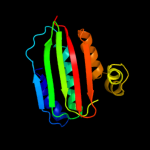
| 18 | d1i58a_ |
|
 |
99.9 |
28 |
Fold:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Superfamily:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Family:Histidine kinase |
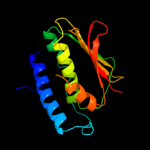
| 19 | c3jz3B_ |
|
 |
99.9 |
26 |
PDB header:transferase
Chain: B: PDB Molecule:sensor protein qsec;
PDBTitle: structure of the cytoplasmic segment of histidine kinase qsec
|
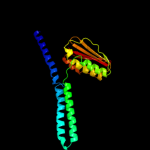
| 20 | c3gieA_ |
|
 |
99.8 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:sensor histidine kinase desk;
PDBTitle: crystal structure of deskc_h188e in complex with amp-pcp
|
| 21 | d2hkja3 |
|
not modelled |
99.8 |
17 |
Fold:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Superfamily:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Family:DNA gyrase/MutL, N-terminal domain |
| 22 | d1r62a_ |
|
not modelled |
99.8 |
22 |
Fold:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Superfamily:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Family:Histidine kinase |
| 23 | c3ehgA_ |
|
not modelled |
99.7 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:sensor kinase (yocf protein);
PDBTitle: crystal structure of the atp-binding domain of desk in complex with2 atp
|
| 24 | c2zbkB_ |
|
not modelled |
99.7 |
20 |
PDB header:isomerase
Chain: B: PDB Molecule:type 2 dna topoisomerase 6 subunit b;
PDBTitle: crystal structure of an intact type ii dna topoisomerase:2 insights into dna transfer mechanisms
|
| 25 | c1mx0D_ |
|
not modelled |
99.7 |
20 |
PDB header:isomerase
Chain: D: PDB Molecule:type ii dna topoisomerase vi subunit b;
PDBTitle: structure of topoisomerase subunit
|
| 26 | c3zxqA_ |
|
not modelled |
99.7 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:hypoxia sensor histidine kinase response regulator dost;
PDBTitle: crystal structure of the atp-binding domain of mycobacterium2 tuberculosis dost
|
| 27 | c3zxoB_ |
|
not modelled |
99.6 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:redox sensor histidine kinase response regulator devs;
PDBTitle: crystal structure of the mutant atp-binding domain of2 mycobacterium tuberculosis doss
|
| 28 | c2q2eB_ |
|
not modelled |
99.6 |
21 |
PDB header:isomerase
Chain: B: PDB Molecule:type 2 dna topoisomerase 6 subunit b;
PDBTitle: crystal structure of the topoisomerase vi holoenzyme from2 methanosarcina mazei
|
| 29 | d1h7sa2 |
|
not modelled |
99.6 |
16 |
Fold:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Superfamily:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Family:DNA gyrase/MutL, N-terminal domain |
| 30 | d1bkna2 |
|
not modelled |
99.5 |
18 |
Fold:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Superfamily:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Family:DNA gyrase/MutL, N-terminal domain |
| 31 | d1th8a_ |
|
not modelled |
99.5 |
20 |
Fold:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Superfamily:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Family:Histidine kinase |
| 32 | d1b63a2 |
|
not modelled |
99.5 |
16 |
Fold:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Superfamily:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Family:DNA gyrase/MutL, N-terminal domain |
| 33 | d1y8oa2 |
|
not modelled |
99.3 |
20 |
Fold:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Superfamily:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Family:alpha-ketoacid dehydrogenase kinase, C-terminal domain |
| 34 | d1ixma_ |
|
not modelled |
99.2 |
9 |
Fold:Sporulation response regulatory protein Spo0B
Superfamily:Sporulation response regulatory protein Spo0B
Family:Sporulation response regulatory protein Spo0B |
| 35 | c3na3A_ |
|
not modelled |
99.0 |
22 |
PDB header:protein binding
Chain: A: PDB Molecule:dna mismatch repair protein mlh1;
PDBTitle: mutl protein homolog 1 isoform 1 from homo sapiens
|
| 36 | c3zrwB_ |
|
not modelled |
99.0 |
61 |
PDB header:signaling protein
Chain: B: PDB Molecule:af1503 protein, osmolarity sensor protein envz;
PDBTitle: the structure of the dimeric hamp-dhp fusion a291v mutant
|
| 37 | c3h4lB_ |
|
not modelled |
98.7 |
19 |
PDB header:dna binding protein, protein binding
Chain: B: PDB Molecule:dna mismatch repair protein pms1;
PDBTitle: crystal structure of n terminal domain of a dna repair protein
|
| 38 | c3lnrA_ |
|
not modelled |
98.5 |
6 |
PDB header:signaling protein
Chain: A: PDB Molecule:aerotaxis transducer aer2;
PDBTitle: crystal structure of poly-hamp domains from the p. aeruginosa soluble2 receptor aer2
|
| 39 | c1bknA_ |
|
not modelled |
98.4 |
19 |
PDB header:dna repair
Chain: A: PDB Molecule:mutl;
PDBTitle: crystal structure of an n-terminal 40kd fragment of e. coli2 dna mismatch repair protein mutl
|
| 40 | d1pvga2 |
|
not modelled |
98.3 |
21 |
Fold:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Superfamily:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Family:DNA gyrase/MutL, N-terminal domain |
| 41 | c1ea6A_ |
|
not modelled |
98.2 |
24 |
PDB header:dna repair
Chain: A: PDB Molecule:pms1 protein homolog 2;
PDBTitle: n-terminal 40kda fragment of nhpms2 complexed with adp
|
| 42 | d1kija2 |
|
not modelled |
98.2 |
19 |
Fold:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Superfamily:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Family:DNA gyrase/MutL, N-terminal domain |
| 43 | d1s16a2 |
|
not modelled |
98.2 |
15 |
Fold:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Superfamily:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Family:DNA gyrase/MutL, N-terminal domain |
| 44 | d1ei1a2 |
|
not modelled |
98.2 |
24 |
Fold:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Superfamily:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Family:DNA gyrase/MutL, N-terminal domain |
| 45 | c1ei1B_ |
|
not modelled |
98.1 |
25 |
PDB header:isomerase
Chain: B: PDB Molecule:dna gyrase b;
PDBTitle: dimerization of e. coli dna gyrase b provides a structural mechanism2 for activating the atpase catalytic center
|
| 46 | c1y4sA_ |
|
not modelled |
98.0 |
20 |
PDB header:chaperone
Chain: A: PDB Molecule:chaperone protein htpg;
PDBTitle: conformation rearrangement of heat shock protein 90 upon2 adp binding
|
| 47 | c1kijB_ |
|
not modelled |
98.0 |
18 |
PDB header:isomerase
Chain: B: PDB Molecule:dna gyrase subunit b;
PDBTitle: crystal structure of the 43k atpase domain of thermus thermophilus2 gyrase b in complex with novobiocin
|
| 48 | c1s16B_ |
|
not modelled |
98.0 |
16 |
PDB header:isomerase
Chain: B: PDB Molecule:topoisomerase iv subunit b;
PDBTitle: crystal structure of e. coli topoisomerase iv pare 43kda subunit2 complexed with adpnp
|
| 49 | d1uyla_ |
|
not modelled |
97.9 |
11 |
Fold:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Superfamily:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Family:Heat shock protein 90, HSP90, N-terminal domain |
| 50 | c3iedA_ |
|
not modelled |
97.9 |
12 |
PDB header:chaperone
Chain: A: PDB Molecule:heat shock protein;
PDBTitle: crystal structure of n-terminal domain of plasmodium falciparum hsp902 (pf14_0417) in complex with amppn
|
| 51 | c2fwyA_ |
|
not modelled |
97.9 |
11 |
PDB header:chaperone
Chain: A: PDB Molecule:heat shock protein hsp 90-alpha;
PDBTitle: structure of human hsp90-alpha bound to the potent water2 soluble inhibitor pu-h64
|
| 52 | c1qzrA_ |
|
not modelled |
97.9 |
27 |
PDB header:isomerase
Chain: A: PDB Molecule:dna topoisomerase ii;
PDBTitle: crystal structure of the atpase region of saccharomyces cerevisiae2 topoisomerase ii bound to icrf-187 (dexrazoxane)
|
| 53 | d2asxa1 |
|
not modelled |
97.8 |
15 |
Fold:HAMP domain-like
Superfamily:HAMP domain-like
Family:HAMP domain |
| 54 | d2iwxa1 |
|
not modelled |
97.8 |
18 |
Fold:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Superfamily:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Family:Heat shock protein 90, HSP90, N-terminal domain |
| 55 | c1zwhA_ |
|
not modelled |
97.8 |
20 |
PDB header:chaperone
Chain: A: PDB Molecule:atp-dependent molecular chaperone hsp82;
PDBTitle: yeast hsp82 in complex with the novel hsp90 inhibitor radester amine
|
| 56 | c3ke6A_ |
|
not modelled |
97.8 |
23 |
PDB header:unknown function
Chain: A: PDB Molecule:protein rv1364c/mt1410;
PDBTitle: the crystal structure of the rsbu and rsbw domains of rv1364c from2 mycobacterium tuberculosis
|
| 57 | c1zxnB_ |
|
not modelled |
97.8 |
21 |
PDB header:isomerase
Chain: B: PDB Molecule:dna topoisomerase ii, alpha isozyme;
PDBTitle: human dna topoisomerase iia atpase/adp
|
| 58 | c2cg9A_ |
|
not modelled |
97.7 |
22 |
PDB header:chaperone
Chain: A: PDB Molecule:atp-dependent molecular chaperone hsp82;
PDBTitle: crystal structure of an hsp90-sba1 closed chaperone complex
|
| 59 | c2iopD_ |
|
not modelled |
97.7 |
22 |
PDB header:chaperone
Chain: D: PDB Molecule:chaperone protein htpg;
PDBTitle: crystal structure of full-length htpg, the escherichia coli2 hsp90, bound to adp
|
| 60 | c2akpA_ |
|
not modelled |
97.7 |
14 |
PDB header:chaperone
Chain: A: PDB Molecule:atp-dependent molecular chaperone hsp82;
PDBTitle: hsp90 delta24-n210 mutant
|
| 61 | c3pehB_ |
|
not modelled |
97.7 |
20 |
PDB header:chaperone
Chain: B: PDB Molecule:endoplasmin homolog;
PDBTitle: crystal structure of the n-terminal domain of an hsp90 from plasmodium2 falciparum, pfl1070c in the presence of a thienopyrimidine derivative
|
| 62 | c2iorA_ |
|
not modelled |
97.6 |
22 |
PDB header:chaperone
Chain: A: PDB Molecule:chaperone protein htpg;
PDBTitle: crystal structure of the n-terminal domain of htpg, the2 escherichia coli hsp90, bound to adp
|
| 63 | d1uyma_ |
|
not modelled |
97.5 |
15 |
Fold:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Superfamily:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Family:Heat shock protein 90, HSP90, N-terminal domain |
| 64 | c3g7bB_ |
|
not modelled |
97.5 |
17 |
PDB header:isomerase
Chain: B: PDB Molecule:dna gyrase subunit b;
PDBTitle: staphylococcus aureus gyrase b co-complex with inhibitor
|
| 65 | d2gqpa1 |
|
not modelled |
97.5 |
16 |
Fold:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Superfamily:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Family:Heat shock protein 90, HSP90, N-terminal domain |
| 66 | d1s14a_ |
|
not modelled |
97.4 |
15 |
Fold:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Superfamily:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Family:DNA gyrase/MutL, N-terminal domain |
| 67 | c2o1wB_ |
|
not modelled |
97.2 |
15 |
PDB header:chaperone
Chain: B: PDB Molecule:endoplasmin;
PDBTitle: structure of n-terminal plus middle domains (n+m) of grp94
|
| 68 | d1qy5a_ |
|
not modelled |
97.1 |
19 |
Fold:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Superfamily:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Family:Heat shock protein 90, HSP90, N-terminal domain |
| 69 | d1joya_ |
|
not modelled |
96.7 |
100 |
Fold:ROP-like
Superfamily:Homodimeric domain of signal transducing histidine kinase
Family:Homodimeric domain of signal transducing histidine kinase |
| 70 | c2o1uA_ |
|
not modelled |
96.1 |
22 |
PDB header:chaperone
Chain: A: PDB Molecule:endoplasmin;
PDBTitle: structure of full length grp94 with amp-pnp bound
|
| 71 | c3lnuA_ |
|
not modelled |
96.0 |
24 |
PDB header:isomerase
Chain: A: PDB Molecule:topoisomerase iv subunit b;
PDBTitle: crystal structure of pare subunit
|
| 72 | c2rm8A_ |
|
not modelled |
94.7 |
15 |
PDB header:signaling protein
Chain: A: PDB Molecule:sensory rhodopsin ii transducer;
PDBTitle: the solution structure of phototactic transducer protein2 htrii linker region from natronomonas pharaonis
|
| 73 | c3cwvB_ |
|
not modelled |
92.3 |
7 |
PDB header:isomerase
Chain: B: PDB Molecule:dna gyrase, b subunit, truncated;
PDBTitle: crystal structure of b-subunit of the dna gyrase from myxococcus2 xanthus
|
| 74 | d2c2aa1 |
|
not modelled |
90.2 |
24 |
Fold:ROP-like
Superfamily:Homodimeric domain of signal transducing histidine kinase
Family:Homodimeric domain of signal transducing histidine kinase |
| 75 | c3lmmA_ |
|
not modelled |
84.3 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of the dip2311 protein from2 corynebacterium diphtheriae, northeast structural genomics3 consortium target cdr35
|
| 76 | d1wmaa1 |
|
not modelled |
78.9 |
30 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 77 | c3s8mA_ |
|
not modelled |
59.3 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:enoyl-acp reductase;
PDBTitle: the crystal structure of fabv
|
| 78 | d1g0oa_ |
|
not modelled |
59.3 |
12 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 79 | c2b4qB_ |
|
not modelled |
54.0 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:rhamnolipids biosynthesis 3-oxoacyl-[acyl-
PDBTitle: pseudomonas aeruginosa rhlg/nadp active-site complex
|
| 80 | d2o23a1 |
|
not modelled |
53.8 |
30 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 81 | d1zema1 |
|
not modelled |
50.6 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 82 | c2z1nA_ |
|
not modelled |
47.0 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dehydrogenase;
PDBTitle: crystal structure of ape0912 from aeropyrum pernix k1
|
| 83 | c3h7aC_ |
|
not modelled |
46.9 |
15 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:short chain dehydrogenase;
PDBTitle: crystal structure of short-chain dehydrogenase from2 rhodopseudomonas palustris
|
| 84 | d1oaaa_ |
|
not modelled |
46.4 |
11 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 85 | c3n0rA_ |
|
not modelled |
45.2 |
9 |
PDB header:signaling protein
Chain: A: PDB Molecule:response regulator;
PDBTitle: structure of the phyr stress response regulator at 1.25 angstrom2 resolution
|
| 86 | c2crqA_ |
|
not modelled |
45.1 |
10 |
PDB header:translation
Chain: A: PDB Molecule:mitochondrial translational initiation factor 3;
PDBTitle: solution structure of c-terminal domain of riken cdna2 2810012l14
|
| 87 | d1spxa_ |
|
not modelled |
44.5 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 88 | d1y5ma1 |
|
not modelled |
43.6 |
12 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 89 | c3ctmH_ |
|
not modelled |
43.5 |
14 |
PDB header:oxidoreductase
Chain: H: PDB Molecule:carbonyl reductase;
PDBTitle: crystal structure of a carbonyl reductase from candida2 parapsilosis with anti-prelog stereo-specificity
|
| 90 | c2p68A_ |
|
not modelled |
41.2 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-oxoacyl-[acyl-carrier-protein] reductase;
PDBTitle: crystal structure of aq_1716 from aquifex aeolicus vf5
|
| 91 | c3o38D_ |
|
not modelled |
39.9 |
21 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:short chain dehydrogenase;
PDBTitle: crystal structure of a short chain dehydrogenase from mycobacterium2 smegmatis
|
| 92 | c3rd5A_ |
|
not modelled |
39.7 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:mypaa.01249.c;
PDBTitle: crystal structure of a putative uncharacterized protein from2 mycobacterium paratuberculosis
|
| 93 | c3i1jB_ |
|
not modelled |
39.7 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase, short chain
PDBTitle: structure of a putative short chain dehydrogenase from2 pseudomonas syringae
|
| 94 | c2jahB_ |
|
not modelled |
38.7 |
12 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:clavulanic acid dehydrogenase;
PDBTitle: biochemical and structural analysis of the clavulanic acid2 dehydeogenase (cad) from streptomyces clavuligerus
|
| 95 | c3imfA_ |
|
not modelled |
38.3 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:short chain dehydrogenase;
PDBTitle: 1.99 angstrom resolution crystal structure of a short chain2 dehydrogenase from bacillus anthracis str. 'ames ancestor'
|
| 96 | c3qivA_ |
|
not modelled |
38.2 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:short-chain dehydrogenase or 3-oxoacyl-[acyl-carrier-
PDBTitle: crystal structure of a putative short-chain dehydrogenase or 3-2 oxoacyl-[acyl-carrier-protein] reductase from mycobacterium3 paratuberculosis atcc baa-968 / k-10
|
| 97 | c3afnC_ |
|
not modelled |
37.9 |
23 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:carbonyl reductase;
PDBTitle: crystal structure of aldose reductase a1-r complexed with nadp
|
| 98 | d1gz6a_ |
|
not modelled |
37.3 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 99 | c3ftpD_ |
|
not modelled |
37.0 |
19 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:3-oxoacyl-[acyl-carrier protein] reductase;
PDBTitle: crystal structure of 3-ketoacyl-(acyl-carrier-protein)2 reductase from burkholderia pseudomallei at 2.05 a3 resolution
|
| 100 | d2bela_ |
|
not modelled |
36.9 |
8 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 101 | c3f5sA_ |
|
not modelled |
36.8 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dehydrogenase;
PDBTitle: crystal structure of putatitve short chain dehydrogenase from shigella2 flexneri 2a str. 301
|
| 102 | d2h7ma1 |
|
not modelled |
36.0 |
12 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 103 | c3sx2F_ |
|
not modelled |
35.9 |
14 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:putative 3-ketoacyl-(acyl-carrier-protein) reductase;
PDBTitle: crystal structure of a putative 3-ketoacyl-(acyl-carrier-protein)2 reductase from mycobacterium paratuberculosis in complex with nad
|
| 104 | c3tfoD_ |
|
not modelled |
35.7 |
18 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:putative 3-oxoacyl-(acyl-carrier-protein) reductase;
PDBTitle: crystal structure of a putative 3-oxoacyl-(acyl-carrier-protein)2 reductase from sinorhizobium meliloti
|
| 105 | c2o2sA_ |
|
not modelled |
35.1 |
28 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:enoyl-acyl carrier reductase;
PDBTitle: the structure of t. gondii enoyl acyl carrier protein reductase in2 complex with nad and triclosan
|
| 106 | c3uf0A_ |
|
not modelled |
34.8 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:short-chain dehydrogenase/reductase sdr;
PDBTitle: crystal structure of a putative nad(p) dependent gluconate 5-2 dehydrogenase from beutenbergia cavernae(efi target efi-502044) with3 bound nadp (low occupancy)
|
| 107 | c3gafF_ |
|
not modelled |
34.7 |
15 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:7-alpha-hydroxysteroid dehydrogenase;
PDBTitle: 2.2a crystal structure of 7-alpha-hydroxysteroid2 dehydrogenase from brucella melitensis
|
| 108 | d1e7wa_ |
|
not modelled |
34.7 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 109 | c2fwmX_ |
|
not modelled |
34.4 |
17 |
PDB header:oxidoreductase
Chain: X: PDB Molecule:2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase;
PDBTitle: crystal structure of e. coli enta, a 2,3-dihydrodihydroxy benzoate2 dehydrogenase
|
| 110 | d1h5qa_ |
|
not modelled |
34.4 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 111 | c3ksuA_ |
|
not modelled |
34.2 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-oxoacyl-acyl carrier protein reductase;
PDBTitle: crystal structure of short-chain dehydrogenase from2 oenococcus oeni psu-1
|
| 112 | c3tjrA_ |
|
not modelled |
34.1 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:short chain dehydrogenase;
PDBTitle: crystal structure of a rv0851c ortholog short chain dehydrogenase from2 mycobacterium paratuberculosis
|
| 113 | c3s55F_ |
|
not modelled |
33.9 |
7 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:putative short-chain dehydrogenase/reductase;
PDBTitle: crystal structure of a putative short-chain dehydrogenase/reductase2 from mycobacterium abscessus bound to nad
|
| 114 | d1yxma1 |
|
not modelled |
33.9 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 115 | c3f9iB_ |
|
not modelled |
33.6 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:3-oxoacyl-[acyl-carrier-protein] reductase;
PDBTitle: crystal structure of 3-ketoacyl-(acyl-carrier-protein) reductase2 rickettsia prowazekii
|
| 116 | c3pxxE_ |
|
not modelled |
33.6 |
10 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:carveol dehydrogenase;
PDBTitle: crystal structure of carveol dehydrogenase from mycobacterium avium2 bound to nicotinamide adenine dinucleotide
|
| 117 | c3ai3A_ |
|
not modelled |
33.3 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadph-sorbose reductase;
PDBTitle: the crystal structure of l-sorbose reductase from gluconobacter2 frateurii complexed with nadph and l-sorbose
|
| 118 | d1xsea_ |
|
not modelled |
33.3 |
12 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 119 | d1odka_ |
|
not modelled |
33.1 |
11 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 120 | c3r1iB_ |
|
not modelled |
33.0 |
12 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:short-chain type dehydrogenase/reductase;
PDBTitle: crystal structure of a short-chain type dehydrogenase/reductase from2 mycobacterium marinum
|

