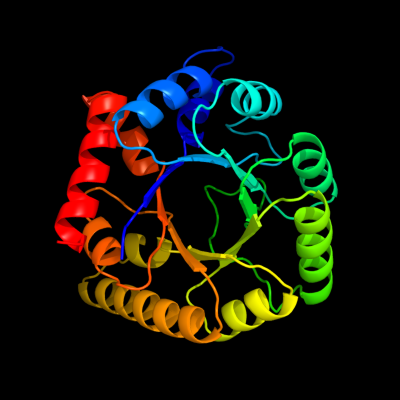
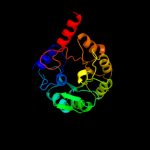
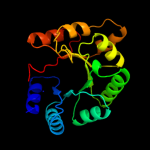
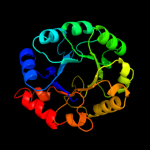
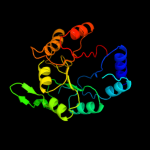
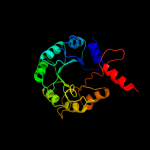
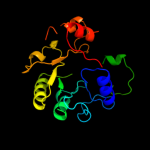
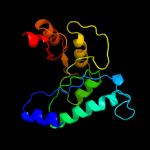
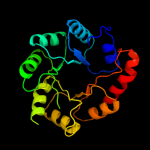
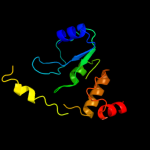
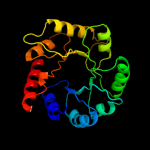
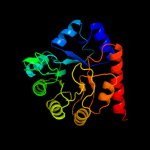
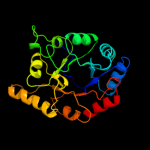
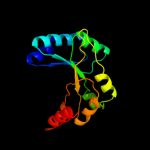
1 d1m5wa_
100.0
100
Fold: TIM beta/alpha-barrelSuperfamily: Pyridoxine 5'-phosphate synthaseFamily: Pyridoxine 5'-phosphate synthase2 c3gk0H_
100.0
67
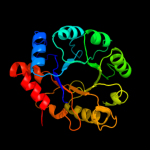
PDB header: transferaseChain: H: PDB Molecule: pyridoxine 5'-phosphate synthase;PDBTitle: crystal structure of pyridoxal phosphate biosynthetic2 protein from burkholderia pseudomallei
3 c3o6cA_
100.0
48
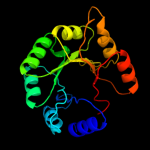
PDB header: transferaseChain: A: PDB Molecule: pyridoxine 5'-phosphate synthase;PDBTitle: pyridoxal phosphate biosynthetic protein pdxj from campylobacter2 jejuni
4 c3igsB_
98.3
16
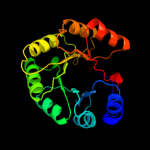
PDB header: isomeraseChain: B: PDB Molecule: n-acetylmannosamine-6-phosphate 2-epimerase 2;PDBTitle: structure of the salmonella enterica n-acetylmannosamine-6-phosphate2 2-epimerase
5 c3qjaA_
97.9
16
PDB header: lyaseChain: A: PDB Molecule: indole-3-glycerol phosphate synthase;PDBTitle: crystal structure of the mycobacterium tuberculosis indole-3-glycerol2 phosphate synthase (trpc) in apo form
6 d2flia1
97.8
13
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: D-ribulose-5-phosphate 3-epimerase7 d1yxya1
97.8
18
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: NanE-like8 c3q58A_
97.6
16
PDB header: isomeraseChain: A: PDB Molecule: n-acetylmannosamine-6-phosphate 2-epimerase;PDBTitle: structure of n-acetylmannosamine-6-phosphate epimerase from salmonella2 enterica
9 c3lotC_
97.4
21
PDB header: structure genomics, unknown functionChain: C: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of protein of unknown function (np_070038.1) from2 archaeoglobus fulgidus at 1.89 a resolution
10 c3gr7A_
97.4
18
PDB header: oxidoreductaseChain: A: PDB Molecule: nadph dehydrogenase;PDBTitle: structure of oye from geobacillus kaustophilus, hexagonal2 crystal form
11 d1xcfa_
97.3
19
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes12 c3e02A_
97.3
20
PDB header: metal binding proteinChain: A: PDB Molecule: uncharacterized protein duf849;PDBTitle: crystal structure of a duf849 family protein (bxe_c0271) from2 burkholderia xenovorans lb400 at 1.90 a resolution
13 d1tqja_
97.3
13
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: D-ribulose-5-phosphate 3-epimerase14 d1vc4a_
97.3
21
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes15 c2c3zA_
97.2
15
PDB header: lyaseChain: A: PDB Molecule: indole-3-glycerol phosphate synthase;PDBTitle: crystal structure of a truncated variant of indole-3-2 glycerol phosphate synthase from sulfolobus solfataricus
16 d1rd5a_
97.1
20
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes17 d1wv2a_
97.1
16
Fold: TIM beta/alpha-barrelSuperfamily: ThiG-likeFamily: ThiG-like18 d1wbha1
97.1
15
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase19 d1q6oa_
97.0
22
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Decarboxylase20 c3inpA_
96.8
15
PDB header: isomeraseChain: A: PDB Molecule: d-ribulose-phosphate 3-epimerase;PDBTitle: 2.05 angstrom resolution crystal structure of d-ribulose-phosphate 3-2 epimerase from francisella tularensis.
21 c3hf3A_
not modelled
96.8
20
PDB header: oxidoreductaseChain: A: PDB Molecule: chromate reductase;PDBTitle: old yellow enzyme from thermus scotoductus sa-01
22 d1rpxa_
not modelled
96.8
13
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: D-ribulose-5-phosphate 3-epimerase23 d1h1ya_
not modelled
96.8
12
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: D-ribulose-5-phosphate 3-epimerase24 c3c6cA_
not modelled
96.7
23
PDB header: hydrolaseChain: A: PDB Molecule: 3-keto-5-aminohexanoate cleavage enzyme;PDBTitle: crystal structure of a putative 3-keto-5-aminohexanoate cleavage2 enzyme (reut_c6226) from ralstonia eutropha jmp134 at 1.72 a3 resolution
25 c3e49A_
not modelled
96.7
25
PDB header: metal binding proteinChain: A: PDB Molecule: uncharacterized protein duf849 with a tim barrel fold;PDBTitle: crystal structure of a prokaryotic domain of unknown function (duf849)2 with a tim barrel fold (bxe_c0966) from burkholderia xenovorans lb4003 at 1.75 a resolution
26 c3ct7E_
not modelled
96.6
14
PDB header: isomeraseChain: E: PDB Molecule: d-allulose-6-phosphate 3-epimerase;PDBTitle: crystal structure of d-allulose 6-phosphate 3-epimerase2 from escherichia coli k-12
27 d1y0ea_
not modelled
96.6
20
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: NanE-like28 d1thfd_
not modelled
96.4
11
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Histidine biosynthesis enzymes29 d1tqxa_
not modelled
96.3
11
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: D-ribulose-5-phosphate 3-epimerase30 c2v82A_
not modelled
96.2
14
PDB header: lyaseChain: A: PDB Molecule: 2-dehydro-3-deoxy-6-phosphogalactonate aldolase;PDBTitle: kdpgal complexed to kdpgal
31 c2htmB_
not modelled
96.1
16
PDB header: biosynthetic proteinChain: B: PDB Molecule: thiazole biosynthesis protein thig;PDBTitle: crystal structure of ttha0676 from thermus thermophilus hb8
32 c3l5aA_
not modelled
95.9
13
PDB header: oxidoreductaseChain: A: PDB Molecule: nadh/flavin oxidoreductase/nadh oxidase;PDBTitle: crystal structure of a probable nadh-dependent flavin oxidoreductase2 from staphylococcus aureus
33 c2y7eA_
not modelled
95.9
19
PDB header: lyaseChain: A: PDB Molecule: 3-keto-5-aminohexanoate cleavage enzyme;PDBTitle: crystal structure of the 3-keto-5-aminohexanoate cleavage enzyme2 (kce) from candidatus cloacamonas acidaminovorans (tetragonal form)
34 d1wa3a1
not modelled
95.9
13
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase35 d1i4na_
not modelled
95.7
14
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes36 c2h90A_
not modelled
95.6
16
PDB header: oxidoreductaseChain: A: PDB Molecule: xenobiotic reductase a;PDBTitle: xenobiotic reductase a in complex with coumarin
37 d1j5ta_
not modelled
95.5
11
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes38 c2bdqA_
not modelled
95.4
19
PDB header: metal transportChain: A: PDB Molecule: copper homeostasis protein cutc;PDBTitle: crystal structure of the putative copper homeostasis2 protein cutc from streptococcus agalactiae, northeast3 strucural genomics target sar15.
39 d1a53a_
not modelled
95.3
16
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes40 d1tv5a1
not modelled
95.2
14
Fold: TIM beta/alpha-barrelSuperfamily: FMN-linked oxidoreductasesFamily: FMN-linked oxidoreductases41 c1tv5A_
not modelled
95.2
14
PDB header: oxidoreductaseChain: A: PDB Molecule: dihydroorotate dehydrogenase homolog, mitochondrial;PDBTitle: plasmodium falciparum dihydroorotate dehydrogenase with a bound2 inhibitor
42 c3chvA_
not modelled
95.1
15
PDB header: metal binding proteinChain: A: PDB Molecule: prokaryotic domain of unknown function (duf849) with a timPDBTitle: crystal structure of a prokaryotic domain of unknown function (duf849)2 member (spoa0042) from silicibacter pomeroyi dss-3 at 1.45 a3 resolution
43 d1piia2
not modelled
95.1
19
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes44 d1vhca_
not modelled
95.1
22
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase45 c3ajxA_
not modelled
95.1
19
PDB header: lyaseChain: A: PDB Molecule: 3-hexulose-6-phosphate synthase;PDBTitle: crystal structure of 3-hexulose-6-phosphate synthase
46 c3jr2D_
not modelled
95.1
20
PDB header: biosynthetic proteinChain: D: PDB Molecule: hexulose-6-phosphate synthase sgbh;PDBTitle: x-ray crystal structure of the mg-bound 3-keto-l-gulonate-6-phosphate2 decarboxylase from vibrio cholerae o1 biovar el tor str. n16961
47 c3f4wA_
not modelled
95.0
14
PDB header: synthase, lyaseChain: A: PDB Molecule: putative hexulose 6 phosphate synthase;PDBTitle: the 1.65a crystal structure of 3-hexulose-6-phosphate2 synthase from salmonella typhimurium
48 d1z41a1
not modelled
94.9
18
Fold: TIM beta/alpha-barrelSuperfamily: FMN-linked oxidoreductasesFamily: FMN-linked oxidoreductases49 c3cixA_
not modelled
94.8
21
PDB header: adomet binding proteinChain: A: PDB Molecule: fefe-hydrogenase maturase;PDBTitle: x-ray structure of the [fefe]-hydrogenase maturase hyde from2 thermotoga maritima in complex with thiocyanate
50 d1q45a_
not modelled
94.7
13
Fold: TIM beta/alpha-barrelSuperfamily: FMN-linked oxidoreductasesFamily: FMN-linked oxidoreductases51 d1bxca_
not modelled
94.7
21
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase52 c3dx5A_
not modelled
94.7
18
PDB header: lyaseChain: A: PDB Molecule: uncharacterized protein asbf;PDBTitle: crystal structure of the probable 3-dhs dehydratase asbf involved in2 the petrobactin synthesis from bacillus anthracis
53 c3thaB_
not modelled
94.5
16
PDB header: lyaseChain: B: PDB Molecule: tryptophan synthase alpha chain;PDBTitle: tryptophan synthase subunit alpha from campylobacter jejuni.
54 d1vyra_
not modelled
94.5
18
Fold: TIM beta/alpha-barrelSuperfamily: FMN-linked oxidoreductasesFamily: FMN-linked oxidoreductases55 d1qt1a_
not modelled
94.5
19
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase56 d1muwa_
not modelled
94.4
18
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase57 c2fptA_
not modelled
94.4
19
PDB header: oxidoreductaseChain: A: PDB Molecule: dihydroorotate dehydrogenase, mitochondrial;PDBTitle: dual binding mode of a novel series of dhodh inhibitors
58 c3qc3B_
not modelled
94.1
13
PDB header: isomeraseChain: B: PDB Molecule: d-ribulose-5-phosphate-3-epimerase;PDBTitle: crystal structure of a d-ribulose-5-phosphate-3-epimerase (np_954699)2 from homo sapiens at 2.20 a resolution
59 c1ps9A_
not modelled
94.0
15
PDB header: oxidoreductaseChain: A: PDB Molecule: 2,4-dienoyl-coa reductase;PDBTitle: the crystal structure and reaction mechanism of e. coli 2,4-2 dienoyl coa reductase
60 d1xima_
not modelled
93.9
22
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase61 c3exsB_
not modelled
93.8
21
PDB header: lyaseChain: B: PDB Molecule: rmpd (hexulose-6-phosphate synthase);PDBTitle: crystal structure of kgpdc from streptococcus mutans in2 complex with d-r5p
62 d1h5ya_
not modelled
93.8
17
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Histidine biosynthesis enzymes63 d1xi3a_
not modelled
93.7
15
Fold: TIM beta/alpha-barrelSuperfamily: Thiamin phosphate synthaseFamily: Thiamin phosphate synthase64 d1f76a_
not modelled
93.7
18
Fold: TIM beta/alpha-barrelSuperfamily: FMN-linked oxidoreductasesFamily: FMN-linked oxidoreductases65 c1yadD_
not modelled
93.5
16
PDB header: transcriptionChain: D: PDB Molecule: regulatory protein teni;PDBTitle: structure of teni from bacillus subtilis
66 d1ka9f_
not modelled
93.5
16
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Histidine biosynthesis enzymes67 d1vhna_
not modelled
93.3
18
Fold: TIM beta/alpha-barrelSuperfamily: FMN-linked oxidoreductasesFamily: FMN-linked oxidoreductases68 c3kruC_
not modelled
93.2
12
PDB header: oxidoreductaseChain: C: PDB Molecule: nadh:flavin oxidoreductase/nadh oxidase;PDBTitle: crystal structure of the thermostable old yellow enzyme from2 thermoanaerobacter pseudethanolicus e39
69 d1r30a_
not modelled
93.2
11
Fold: TIM beta/alpha-barrelSuperfamily: Radical SAM enzymesFamily: Biotin synthase70 c1r30A_
not modelled
93.2
11
PDB header: transferaseChain: A: PDB Molecule: biotin synthase;PDBTitle: the crystal structure of biotin synthase, an s-2 adenosylmethionine-dependent radical enzyme
71 d1djqa1
not modelled
93.2
17
Fold: TIM beta/alpha-barrelSuperfamily: FMN-linked oxidoreductasesFamily: FMN-linked oxidoreductases72 c3no5C_
not modelled
93.2
20
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a pfam duf849 domain containing protein2 (reut_a1631) from ralstonia eutropha jmp134 at 1.90 a resolution
73 c3gkaB_
not modelled
93.2
31
PDB header: oxidoreductaseChain: B: PDB Molecule: n-ethylmaleimide reductase;PDBTitle: crystal structure of n-ethylmaleimidine reductase from2 burkholderia pseudomallei
74 c1xrsB_
not modelled
93.0
19
PDB header: isomeraseChain: B: PDB Molecule: d-lysine 5,6-aminomutase beta subunit;PDBTitle: crystal structure of lysine 5,6-aminomutase in complex with plp,2 cobalamin, and 5'-deoxyadenosine
75 c3qxbB_
not modelled
93.0
24
PDB header: isomeraseChain: B: PDB Molecule: putative xylose isomerase;PDBTitle: crystal structure of a putative xylose isomerase (yp_426450.1) from2 rhodospirillum rubrum atcc 11170 at 1.90 a resolution
76 c2yw3E_
not modelled
92.5
14
PDB header: lyaseChain: E: PDB Molecule: 4-hydroxy-2-oxoglutarate aldolase/2-deydro-3-PDBTitle: crystal structure analysis of the 4-hydroxy-2-oxoglutarate aldolase/2-2 deydro-3-deoxyphosphogluconate aldolase from tthb1
77 d1ps9a1
not modelled
92.1
15
Fold: TIM beta/alpha-barrelSuperfamily: FMN-linked oxidoreductasesFamily: FMN-linked oxidoreductases78 d1i60a_
not modelled
91.9
11
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: IolI-like79 c2gq8A_
not modelled
91.9
20
PDB header: oxidoreductaseChain: A: PDB Molecule: oxidoreductase, fmn-binding;PDBTitle: structure of sye1, an oye homologue from s. ondeidensis, in complex2 with p-hydroxyacetophenone
80 d1uuma_
not modelled
91.8
18
Fold: TIM beta/alpha-barrelSuperfamily: FMN-linked oxidoreductasesFamily: FMN-linked oxidoreductases81 c3aamA_
not modelled
91.8
20
PDB header: hydrolaseChain: A: PDB Molecule: endonuclease iv;PDBTitle: crystal structure of endonuclease iv from thermus thermophilus hb8
82 d1qf6a1
not modelled
91.8
18
Fold: Anticodon-binding domain-likeSuperfamily: Class II aaRS ABD-relatedFamily: Anticodon-binding domain of Class II aaRS83 d1bxba_
not modelled
91.6
17
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase84 c2ou4C_
not modelled
91.6
17
PDB header: isomeraseChain: C: PDB Molecule: d-tagatose 3-epimerase;PDBTitle: crystal structure of d-tagatose 3-epimerase from2 pseudomonas cichorii
85 d1icpa_
not modelled
91.6
20
Fold: TIM beta/alpha-barrelSuperfamily: FMN-linked oxidoreductasesFamily: FMN-linked oxidoreductases86 c2ekcA_
not modelled
91.3
16
PDB header: lyaseChain: A: PDB Molecule: tryptophan synthase alpha chain;PDBTitle: structural study of project id aq_1548 from aquifex aeolicus vf5
87 d1xm3a_
not modelled
91.0
17
Fold: TIM beta/alpha-barrelSuperfamily: ThiG-likeFamily: ThiG-like88 c2zdsB_
not modelled
90.8
13
PDB header: dna binding proteinChain: B: PDB Molecule: putative dna-binding protein;PDBTitle: crystal structure of sco6571 from streptomyces coelicolor2 a3(2)
89 c1djnB_
not modelled
90.7
19
PDB header: oxidoreductaseChain: B: PDB Molecule: trimethylamine dehydrogenase;PDBTitle: structural and biochemical characterization of recombinant wild type2 trimethylamine dehydrogenase from methylophilus methylotrophus (sp.3 w3a1)
90 d1wx0a1
not modelled
90.5
27
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase91 c3o63B_
not modelled
90.4
20
PDB header: transferaseChain: B: PDB Molecule: probable thiamine-phosphate pyrophosphorylase;PDBTitle: crystal structure of thiamin phosphate synthase from mycobacterium2 tuberculosis
92 d1gwja_
not modelled
90.4
15
Fold: TIM beta/alpha-barrelSuperfamily: FMN-linked oxidoreductasesFamily: FMN-linked oxidoreductases93 c3khjE_
not modelled
90.3
15
PDB header: oxidoreductaseChain: E: PDB Molecule: inosine-5-monophosphate dehydrogenase;PDBTitle: c. parvum inosine monophosphate dehydrogenase bound by inhibitor c64
94 d1yx1a1
not modelled
90.2
18
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: KguE-like95 d1k77a_
not modelled
89.8
13
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Hypothetical protein YgbM (EC1530)96 d1vd6a1
not modelled
89.3
12
Fold: TIM beta/alpha-barrelSuperfamily: PLC-like phosphodiesterasesFamily: Glycerophosphoryl diester phosphodiesterase97 c3kwsB_
not modelled
89.2
23
PDB header: isomeraseChain: B: PDB Molecule: putative sugar isomerase;PDBTitle: crystal structure of putative sugar isomerase (yp_001305149.1) from2 parabacteroides distasonis atcc 8503 at 1.68 a resolution
98 c3s1vD_
not modelled
88.5
17
PDB header: transferaseChain: D: PDB Molecule: probable transaldolase;PDBTitle: transaldolase from thermoplasma acidophilum in complex with d-fructose2 6-phosphate schiff-base intermediate
99 c2ftpA_
not modelled
88.2
9
PDB header: lyaseChain: A: PDB Molecule: hydroxymethylglutaryl-coa lyase;PDBTitle: crystal structure of hydroxymethylglutaryl-coa lyase from pseudomonas2 aeruginosa
100 c2i2xD_
not modelled
88.1
17
PDB header: transferaseChain: D: PDB Molecule: methyltransferase 1;PDBTitle: crystal structure of methanol:cobalamin methyltransferase complex2 mtabc from methanosarcina barkeri
101 d1xrsb1
not modelled
87.8
17
Fold: Flavodoxin-likeSuperfamily: Cobalamin (vitamin B12)-binding domainFamily: Cobalamin (vitamin B12)-binding domain102 d1un7a2
not modelled
87.8
25
Fold: TIM beta/alpha-barrelSuperfamily: Metallo-dependent hydrolasesFamily: N-acetylglucosamine-6-phosphate deacetylase, NagA, catalytic domain103 d1oyaa_
not modelled
87.8
18
Fold: TIM beta/alpha-barrelSuperfamily: FMN-linked oxidoreductasesFamily: FMN-linked oxidoreductases104 d1zfja1
not modelled
87.8
16
Fold: TIM beta/alpha-barrelSuperfamily: Inosine monophosphate dehydrogenase (IMPDH)Family: Inosine monophosphate dehydrogenase (IMPDH)105 d2g0wa1
not modelled
87.7
15
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: IolI-like106 c2x7vA_
not modelled
87.7
10
PDB header: hydrolaseChain: A: PDB Molecule: probable endonuclease 4;PDBTitle: crystal structure of thermotoga maritima endonuclease iv in2 the presence of zinc
107 c3k30B_
not modelled
87.6
18
PDB header: oxidoreductaseChain: B: PDB Molecule: histamine dehydrogenase;PDBTitle: histamine dehydrogenase from nocardiodes simplex
108 c3cqkB_
not modelled
87.5
16
PDB header: isomeraseChain: B: PDB Molecule: l-ribulose-5-phosphate 3-epimerase ulae;PDBTitle: crystal structure of l-xylulose-5-phosphate 3-epimerase ulae (form b)2 complex with zn2+ and sulfate
109 d1vjia_
not modelled
87.5
15
Fold: TIM beta/alpha-barrelSuperfamily: FMN-linked oxidoreductasesFamily: FMN-linked oxidoreductases110 d1mxsa_
not modelled
87.5
16
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase111 d1d3ga_
not modelled
87.3
14
Fold: TIM beta/alpha-barrelSuperfamily: FMN-linked oxidoreductasesFamily: FMN-linked oxidoreductases112 c2yciX_
not modelled
87.0
15
PDB header: transferaseChain: X: PDB Molecule: 5-methyltetrahydrofolate corrinoid/iron sulfur proteinPDBTitle: methyltransferase native
113 d1nyra1
not modelled
86.5
21
Fold: Anticodon-binding domain-likeSuperfamily: Class II aaRS ABD-relatedFamily: Anticodon-binding domain of Class II aaRS114 c2v5jB_
not modelled
86.5
19
PDB header: lyaseChain: B: PDB Molecule: 2,4-dihydroxyhept-2-ene-1,7-dioic acid aldolase;PDBTitle: apo class ii aldolase hpch
115 c3pajA_
not modelled
86.4
16
PDB header: transferaseChain: A: PDB Molecule: nicotinate-nucleotide pyrophosphorylase, carboxylating;PDBTitle: 2.00 angstrom resolution crystal structure of a quinolinate2 phosphoribosyltransferase from vibrio cholerae o1 biovar eltor str.3 n16961
116 c1qapA_
not modelled
86.2
13
PDB header: glycosyltransferaseChain: A: PDB Molecule: quinolinic acid phosphoribosyltransferase;PDBTitle: quinolinic acid phosphoribosyltransferase with bound2 quinolinic acid
117 d1qopa_
not modelled
86.2
18
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes118 d1hc7a1
not modelled
86.2
20
Fold: Anticodon-binding domain-likeSuperfamily: Class II aaRS ABD-relatedFamily: Anticodon-binding domain of Class II aaRS119 c2zvrA_
not modelled
85.9
13
PDB header: isomeraseChain: A: PDB Molecule: uncharacterized protein tm_0416;PDBTitle: crystal structure of a d-tagatose 3-epimerase-related2 protein from thermotoga maritima
120 c2kzhA_
not modelled
85.8
18
PDB header: isomeraseChain: A: PDB Molecule: tryptophan biosynthesis protein trpcf;PDBTitle: three-dimensional structure of a truncated phosphoribosylanthranilate2 isomerase (residues 255-384) from escherichia coli