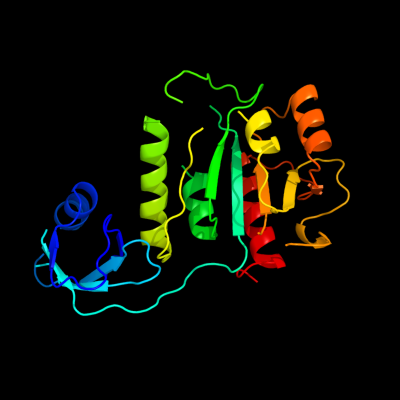
| 1 | c1vhyB_
|
|
 |
100.0 |
63 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical protein hi0303;
PDBTitle: crystal structure of haemophilus influenzae protein hi0303, pfam2 duf558
|
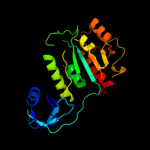
| 2 | c1vhkA_
|
|
 |
100.0 |
28 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein yqeu;
PDBTitle: crystal structure of an hypothetical protein
|
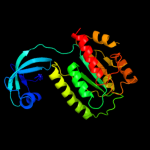
| 3 | c3kw2A_
|
|
 |
100.0 |
30 |
PDB header:transferase
Chain: A: PDB Molecule:probable r-rna methyltransferase;
PDBTitle: crystal structure of probable rrna-methyltransferase from2 porphyromonas gingivalis
|
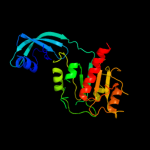
| 4 | c2egwB_
|
|
 |
100.0 |
25 |
PDB header:rna methyltransferase
Chain: B: PDB Molecule:upf0088 protein aq_165;
PDBTitle: crystal structure of rrna methyltransferase with sah ligand
|
| 5 | c1z85B_
|
|
 |
100.0 |
25 |
PDB header:transferase
Chain: B: PDB Molecule:hypothetical protein tm1380;
PDBTitle: crystal structure of a predicted rna methyltransferase (tm1380) from2 thermotoga maritima msb8 at 2.12 a resolution
|
| 6 | d1nxza2
|
|
 |
100.0 |
73 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:YggJ C-terminal domain-like |
| 7 | c2cx8B_
|
|
 |
100.0 |
31 |
PDB header:transferase
Chain: B: PDB Molecule:methyl transferase;
PDBTitle: crystal structure of methyltransferase with ligand(sah)
|
| 8 | d1vhka2
|
|
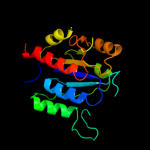 |
100.0 |
33 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:YggJ C-terminal domain-like |
| 9 | d1v6za2
|
|
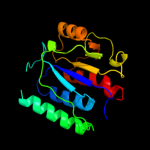 |
100.0 |
31 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:YggJ C-terminal domain-like |
| 10 | d1nxza1
|
|
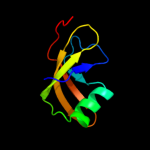 |
99.7 |
51 |
Fold:PUA domain-like
Superfamily:PUA domain-like
Family:YggJ N-terminal domain-like |
| 11 | d1vhka1
|
|
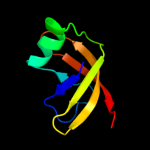 |
99.5 |
19 |
Fold:PUA domain-like
Superfamily:PUA domain-like
Family:YggJ N-terminal domain-like |
| 12 | d2qmma1
|
|
 |
96.4 |
20 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:AF1056-like |
| 13 | c3ai9X_
|
|
 |
96.2 |
16 |
PDB header:transferase
Chain: X: PDB Molecule:upf0217 protein mj1640;
PDBTitle: crystal structure of duf358 protein reveals a putative spout-class2 rrna methyltransferase
|
| 14 | d1v6za1
|
|
 |
95.9 |
28 |
Fold:PUA domain-like
Superfamily:PUA domain-like
Family:YggJ N-terminal domain-like |
| 15 | c1k3rA_
|
|
 |
93.9 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:conserved protein mt0001;
PDBTitle: crystal structure of the methyltransferase with a knot from2 methanobacterium thermoautotrophicum
|
| 16 | d1gz0a1
|
|
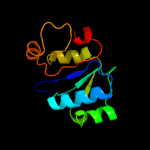 |
93.8 |
13 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:SpoU-like RNA 2'-O ribose methyltransferase |
| 17 | d1ipaa1
|
|
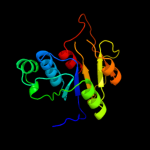 |
93.6 |
14 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:SpoU-like RNA 2'-O ribose methyltransferase |
| 18 | c1gz0G_
|
|
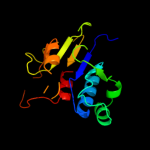 |
93.2 |
11 |
PDB header:transferase
Chain: G: PDB Molecule:hypothetical trna/rrna methyltransferase yjfh;
PDBTitle: 23s ribosomal rna g2251 2'o-methyltransferase rlmb
|
| 19 | c1x7pB_
|
|
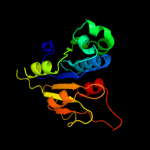 |
92.9 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:rrna methyltransferase;
PDBTitle: crystal structure of the spou methyltransferase avirb from2 streptomyces viridochromogenes in complex with the cofactor adomet
|
| 20 | c1zjrA_
|
|
 |
92.7 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:trna (guanosine-2'-o-)-methyltransferase;
PDBTitle: crystal structure of a. aeolicus trmh/spou trna modifying enzyme
|
| 21 | c2ha8A_ |
|
not modelled |
92.5 |
13 |
PDB header:rna binding protein
Chain: A: PDB Molecule:tar (hiv-1) rna loop binding protein;
PDBTitle: methyltransferase domain of human tar (hiv-1) rna binding2 protein 1
|
| 22 | c2h6rG_ |
|
not modelled |
91.9 |
15 |
PDB header:isomerase
Chain: G: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase (tim) from2 methanocaldococcus jannaschii
|
| 23 | c3ktyA_ |
|
not modelled |
91.6 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:probable methyltransferase;
PDBTitle: crystal structure of probable methyltransferase from bordetella2 pertussis tohama i
|
| 24 | c2i6dA_ |
|
not modelled |
91.0 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:rna methyltransferase, trmh family;
PDBTitle: the structure of a putative rna methyltransferase of the trmh family2 from porphyromonas gingivalis.
|
| 25 | c3onpA_ |
|
not modelled |
90.3 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:trna/rrna methyltransferase (spou);
PDBTitle: crystal structure of trna/rrna methyltransferase spou from rhodobacter2 sphaeroides
|
| 26 | c3l8uA_ |
|
not modelled |
89.1 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:putative rrna methylase;
PDBTitle: crystal structure of smu.1707c, a putative rrna methyltransferase from2 streptococcus mutans ua159
|
| 27 | c3ilkB_ |
|
not modelled |
88.5 |
11 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized trna/rrna methyltransferase hi0380;
PDBTitle: the structure of a probable methylase family protein from haemophilus2 influenzae rd kw20
|
| 28 | d1mxia_ |
|
not modelled |
87.6 |
13 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:SpoU-like RNA 2'-O ribose methyltransferase |
| 29 | d1v2xa_ |
|
not modelled |
87.4 |
15 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:SpoU-like RNA 2'-O ribose methyltransferase |
| 30 | c3ic6A_ |
|
not modelled |
86.0 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative methylase family protein;
PDBTitle: crystal structure of putative methylase family protein from neisseria2 gonorrhoeae
|
| 31 | d1mxsa_ |
|
not modelled |
83.1 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 32 | d1wbha1 |
|
not modelled |
82.6 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 33 | d1k3ra2 |
|
not modelled |
82.2 |
19 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:Hypothetical protein MTH1 (MT0001), dimerisation domain |
| 34 | c1ipaA_ |
|
not modelled |
80.2 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:rna 2'-o-ribose methyltransferase;
PDBTitle: crystal structure of rna 2'-o ribose methyltransferase
|
| 35 | d1rpxa_ |
|
not modelled |
77.9 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
| 36 | c2qiwA_ |
|
not modelled |
76.6 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:pep phosphonomutase;
PDBTitle: crystal structure of a putative phosphoenolpyruvate phosphonomutase2 (ncgl1015, cgl1060) from corynebacterium glutamicum atcc 13032 at3 1.80 a resolution
|
| 37 | c2c3zA_ |
|
not modelled |
75.7 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:indole-3-glycerol phosphate synthase;
PDBTitle: crystal structure of a truncated variant of indole-3-2 glycerol phosphate synthase from sulfolobus solfataricus
|
| 38 | d2flia1 |
|
not modelled |
70.6 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
| 39 | c2v82A_ |
|
not modelled |
68.7 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:2-dehydro-3-deoxy-6-phosphogalactonate aldolase;
PDBTitle: kdpgal complexed to kdpgal
|
| 40 | d1a53a_ |
|
not modelled |
67.9 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 41 | c3inpA_ |
|
not modelled |
67.4 |
13 |
PDB header:isomerase
Chain: A: PDB Molecule:d-ribulose-phosphate 3-epimerase;
PDBTitle: 2.05 angstrom resolution crystal structure of d-ribulose-phosphate 3-2 epimerase from francisella tularensis.
|
| 42 | c3gyqB_ |
|
not modelled |
67.3 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:rrna (adenosine-2'-o-)-methyltransferase;
PDBTitle: structure of the thiostrepton-resistance methyltransferase2 s-adenosyl-l-methionine complex
|
| 43 | d1j5ta_ |
|
not modelled |
67.1 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 44 | c3qjaA_ |
|
not modelled |
66.1 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:indole-3-glycerol phosphate synthase;
PDBTitle: crystal structure of the mycobacterium tuberculosis indole-3-glycerol2 phosphate synthase (trpc) in apo form
|
| 45 | d1vhca_ |
|
not modelled |
65.0 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 46 | c3db3A_ |
|
not modelled |
64.1 |
8 |
PDB header:ligase
Chain: A: PDB Molecule:e3 ubiquitin-protein ligase uhrf1;
PDBTitle: crystal structure of the tandem tudor domains of the e3 ubiquitin-2 protein ligase uhrf1 in complex with trimethylated histone h3-k93 peptide
|
| 47 | c3e5yB_ |
|
not modelled |
63.4 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:trmh family rna methyltransferase;
PDBTitle: crystal structure of trmh family rna methyltransferase from2 burkholderia pseudomallei
|
| 48 | d1znna1 |
|
not modelled |
63.3 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:PdxS-like |
| 49 | c3igsB_ |
|
not modelled |
60.2 |
16 |
PDB header:isomerase
Chain: B: PDB Molecule:n-acetylmannosamine-6-phosphate 2-epimerase 2;
PDBTitle: structure of the salmonella enterica n-acetylmannosamine-6-phosphate2 2-epimerase
|
| 50 | c1znnF_ |
|
not modelled |
59.7 |
14 |
PDB header:biosynthetic protein
Chain: F: PDB Molecule:plp synthase;
PDBTitle: structure of the synthase subunit of plp synthase
|
| 51 | c2e6zA_ |
|
not modelled |
53.2 |
15 |
PDB header:transcription
Chain: A: PDB Molecule:transcription elongation factor spt5;
PDBTitle: solution structure of the second kow motif of human2 transcription elongation factor spt5
|
| 52 | d1piia2 |
|
not modelled |
49.7 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 53 | d1ns5a_ |
|
not modelled |
48.5 |
21 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:YbeA-like |
| 54 | c2e5pA_ |
|
not modelled |
47.5 |
14 |
PDB header:transcription
Chain: A: PDB Molecule:phd finger protein 1;
PDBTitle: solution structure of the tudor domain of phd finger2 protein 1 (phf1 protein)
|
| 55 | c2eqmA_ |
|
not modelled |
46.4 |
15 |
PDB header:transcription
Chain: A: PDB Molecule:phd finger protein 20-like 1;
PDBTitle: solution structure of the tudor domain of phd finger2 protein 20-like 1 [homo sapiens]
|
| 56 | d1y71a1 |
|
not modelled |
45.7 |
20 |
Fold:SH3-like barrel
Superfamily:Kinase-associated protein B-like
Family:Kinase-associated protein B-like |
| 57 | d1i4na_ |
|
not modelled |
44.6 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 58 | d1wa3a1 |
|
not modelled |
44.6 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 59 | c3iz5Y_ |
|
not modelled |
43.7 |
13 |
PDB header:ribosome
Chain: Y: PDB Molecule:60s ribosomal protein l26 (l24p);
PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
|
| 60 | d1to0a_ |
|
not modelled |
43.0 |
13 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:YbeA-like |
| 61 | d2csua1 |
|
not modelled |
39.7 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
| 62 | d3bbda1 |
|
not modelled |
39.7 |
17 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:EMG1/NEP1-like |
| 63 | d1vc4a_ |
|
not modelled |
38.8 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 64 | d1sf8a_ |
|
not modelled |
38.5 |
6 |
Fold:HSP90 C-terminal domain
Superfamily:HSP90 C-terminal domain
Family:HSP90 C-terminal domain |
| 65 | c2equA_ |
|
not modelled |
38.5 |
15 |
PDB header:protein binding
Chain: A: PDB Molecule:phd finger protein 20-like 1;
PDBTitle: solution structure of the tudor domain of phd finger2 protein 20-like 1
|
| 66 | c1yadD_ |
|
not modelled |
38.2 |
12 |
PDB header:transcription
Chain: D: PDB Molecule:regulatory protein teni;
PDBTitle: structure of teni from bacillus subtilis
|
| 67 | c2eqjA_ |
|
not modelled |
37.9 |
14 |
PDB header:transcription
Chain: A: PDB Molecule:metal-response element-binding transcription
PDBTitle: solution structure of the tudor domain of metal-response2 element-binding transcription factor 2
|
| 68 | d1d9ea_ |
|
not modelled |
37.3 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I DAHP synthetase |
| 69 | c2csuB_ |
|
not modelled |
37.3 |
14 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:457aa long hypothetical protein;
PDBTitle: crystal structure of ph0766 from pyrococcus horikoshii ot3
|
| 70 | d1w0ma_ |
|
not modelled |
36.1 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 71 | c2jvvA_ |
|
not modelled |
35.8 |
24 |
PDB header:transcription
Chain: A: PDB Molecule:transcription antitermination protein nusg;
PDBTitle: solution structure of e. coli nusg carboxyterminal domain
|
| 72 | c2kvqG_ |
|
not modelled |
35.8 |
24 |
PDB header:transcription
Chain: G: PDB Molecule:transcription antitermination protein nusg;
PDBTitle: solution structure of nuse:nusg-ctd complex
|
| 73 | d1xi3a_ |
|
not modelled |
35.8 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Thiamin phosphate synthase
Family:Thiamin phosphate synthase |
| 74 | d1te7a_ |
|
not modelled |
35.8 |
12 |
Fold:PUA domain-like
Superfamily:PUA domain-like
Family:yqfB-like |
| 75 | c2yw3E_ |
|
not modelled |
35.7 |
19 |
PDB header:lyase
Chain: E: PDB Molecule:4-hydroxy-2-oxoglutarate aldolase/2-deydro-3-
PDBTitle: crystal structure analysis of the 4-hydroxy-2-oxoglutarate aldolase/2-2 deydro-3-deoxyphosphogluconate aldolase from tthb1
|
| 76 | d1q6oa_ |
|
not modelled |
35.6 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Decarboxylase |
| 77 | c2zpaB_ |
|
not modelled |
35.1 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:uncharacterized protein ypfi;
PDBTitle: crystal structure of trna(met) cytidine acetyltransferase
|
| 78 | c3sz8D_ |
|
not modelled |
34.9 |
9 |
PDB header:transferase
Chain: D: PDB Molecule:2-dehydro-3-deoxyphosphooctonate aldolase 2;
PDBTitle: crystal structure of 2-dehydro-3-deoxyphosphooctonate aldolase from2 burkholderia pseudomallei
|
| 79 | c3qiiA_ |
|
not modelled |
34.0 |
11 |
PDB header:transcription regulator
Chain: A: PDB Molecule:phd finger protein 20;
PDBTitle: crystal structure of tudor domain 2 of human phd finger protein 20
|
| 80 | d1tqja_ |
|
not modelled |
33.4 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
| 81 | d1xm3a_ |
|
not modelled |
33.3 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:ThiG-like
Family:ThiG-like |
| 82 | c2xk0A_ |
|
not modelled |
33.3 |
3 |
PDB header:transcription
Chain: A: PDB Molecule:polycomb protein pcl;
PDBTitle: solution structure of the tudor domain from drosophila2 polycomblike (pcl)
|
| 83 | c3qc3B_ |
|
not modelled |
32.5 |
15 |
PDB header:isomerase
Chain: B: PDB Molecule:d-ribulose-5-phosphate-3-epimerase;
PDBTitle: crystal structure of a d-ribulose-5-phosphate-3-epimerase (np_954699)2 from homo sapiens at 2.20 a resolution
|
| 84 | d1nz9a_ |
|
not modelled |
32.1 |
21 |
Fold:SH3-like barrel
Superfamily:Translation proteins SH3-like domain
Family:N-utilization substance G protein NusG, C-terminal domain |
| 85 | d1vr4a1 |
|
not modelled |
30.6 |
14 |
Fold:Dodecin subunit-like
Superfamily:YbjQ-like
Family:YbjQ-like |
| 86 | c3ih1A_ |
|
not modelled |
28.9 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:methylisocitrate lyase;
PDBTitle: crystal structure of carboxyvinyl-carboxyphosphonate phosphorylmutase2 from bacillus anthracis
|
| 87 | d1vh0a_ |
|
not modelled |
28.9 |
10 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:YbeA-like |
| 88 | d1vqot1 |
|
not modelled |
28.6 |
13 |
Fold:SH3-like barrel
Superfamily:Translation proteins SH3-like domain
Family:Ribosomal proteins L24p and L21e |
| 89 | d2do3a1 |
|
not modelled |
28.3 |
13 |
Fold:SH3-like barrel
Superfamily:Translation proteins SH3-like domain
Family:SPT5 KOW domain-like |
| 90 | c2qjhH_ |
|
not modelled |
27.0 |
12 |
PDB header:lyase
Chain: H: PDB Molecule:putative aldolase mj0400;
PDBTitle: m. jannaschii adh synthase covalently bound to2 dihydroxyacetone phosphate
|
| 91 | c3o63B_ |
|
not modelled |
27.0 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:probable thiamine-phosphate pyrophosphorylase;
PDBTitle: crystal structure of thiamin phosphate synthase from mycobacterium2 tuberculosis
|
| 92 | c3dx5A_ |
|
not modelled |
27.0 |
10 |
PDB header:lyase
Chain: A: PDB Molecule:uncharacterized protein asbf;
PDBTitle: crystal structure of the probable 3-dhs dehydratase asbf involved in2 the petrobactin synthesis from bacillus anthracis
|
| 93 | d1nppa2 |
|
not modelled |
27.0 |
15 |
Fold:SH3-like barrel
Superfamily:Translation proteins SH3-like domain
Family:N-utilization substance G protein NusG, C-terminal domain |
| 94 | d1wjsa_ |
|
not modelled |
26.6 |
9 |
Fold:SH3-like barrel
Superfamily:Tudor/PWWP/MBT
Family:MBT repeat |
| 95 | c3ajxA_ |
|
not modelled |
26.4 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:3-hexulose-6-phosphate synthase;
PDBTitle: crystal structure of 3-hexulose-6-phosphate synthase
|
| 96 | d2tpsa_ |
|
not modelled |
26.4 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Thiamin phosphate synthase
Family:Thiamin phosphate synthase |
| 97 | d1y2ia_ |
|
not modelled |
26.0 |
17 |
Fold:Dodecin subunit-like
Superfamily:YbjQ-like
Family:YbjQ-like |
| 98 | c1y2iC_ |
|
not modelled |
26.0 |
17 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:hypothetical protein s0862;
PDBTitle: crystal structure of mcsg target apc27401 from shigella2 flexneri
|
| 99 | c3navB_ |
|
not modelled |
26.0 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of an alpha subunit of tryptophan synthase from2 vibrio cholerae o1 biovar el tor str. n16961
|
| 100 | d1i60a_ |
|
not modelled |
25.7 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:IolI-like |
| 101 | d2zjrr1 |
|
not modelled |
25.3 |
17 |
Fold:SH3-like barrel
Superfamily:Translation proteins SH3-like domain
Family:Ribosomal proteins L24p and L21e |
| 102 | c3qkbB_ |
|
not modelled |
25.1 |
20 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a protein with unknown function which belongs to2 pfam duf74 family (pepe_0654) from pediococcus pentosaceus atcc 257453 at 2.73 a resolution
|
| 103 | c3exsB_ |
|
not modelled |
24.7 |
11 |
PDB header:lyase
Chain: B: PDB Molecule:rmpd (hexulose-6-phosphate synthase);
PDBTitle: crystal structure of kgpdc from streptococcus mutans in2 complex with d-r5p
|
| 104 | c3q1jA_ |
|
not modelled |
24.7 |
17 |
PDB header:transcription
Chain: A: PDB Molecule:phd finger protein 20;
PDBTitle: crystal structure of tudor domain 1 of human phd finger protein 20
|
| 105 | c3p8bB_ |
|
not modelled |
24.1 |
22 |
PDB header:transferase/transcription
Chain: B: PDB Molecule:transcription antitermination protein nusg;
PDBTitle: x-ray crystal structure of pyrococcus furiosus transcription2 elongation factor spt4/5
|
| 106 | c3f4wA_ |
|
not modelled |
23.9 |
18 |
PDB header:synthase, lyase
Chain: A: PDB Molecule:putative hexulose 6 phosphate synthase;
PDBTitle: the 1.65a crystal structure of 3-hexulose-6-phosphate2 synthase from salmonella typhimurium
|
| 107 | c2xhcA_ |
|
not modelled |
23.8 |
15 |
PDB header:transcription
Chain: A: PDB Molecule:transcription antitermination protein nusg;
PDBTitle: crystal structure of thermotoga maritima n-utilization substance g2 (nusg)
|
| 108 | d1o54a_ |
|
not modelled |
23.3 |
24 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:tRNA(1-methyladenosine) methyltransferase-like |
| 109 | d1wgsa_ |
|
not modelled |
22.8 |
13 |
Fold:SH3-like barrel
Superfamily:Chromo domain-like
Family:Chromo barrel domain |
| 110 | d1wjqa_ |
|
not modelled |
22.7 |
16 |
Fold:SH3-like barrel
Superfamily:Tudor/PWWP/MBT
Family:MBT repeat |
| 111 | c3lyeA_ |
|
not modelled |
22.5 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:oxaloacetate acetyl hydrolase;
PDBTitle: crystal structure of oxaloacetate acetylhydrolase
|
| 112 | d2d59a1 |
|
not modelled |
22.5 |
12 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
| 113 | c2kw8A_ |
|
not modelled |
22.4 |
20 |
PDB header:protein binding
Chain: A: PDB Molecule:lpxtg-site transpeptidase family protein;
PDBTitle: solution structure of bacillus anthracis sortase a (srta)2 transpeptidase
|
| 114 | c3q58A_ |
|
not modelled |
22.3 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:n-acetylmannosamine-6-phosphate 2-epimerase;
PDBTitle: structure of n-acetylmannosamine-6-phosphate epimerase from salmonella2 enterica
|
| 115 | d2nu7a1 |
|
not modelled |
22.2 |
7 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
| 116 | c2e5qA_ |
|
not modelled |
22.0 |
15 |
PDB header:transcription
Chain: A: PDB Molecule:phd finger protein 19;
PDBTitle: solution structure of the tudor domain of phd finger2 protein 19, isoform b [homo sapiens]
|
| 117 | d1o6da_ |
|
not modelled |
21.5 |
18 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:YbeA-like |
| 118 | d1o60a_ |
|
not modelled |
21.4 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I DAHP synthetase |
| 119 | c2rd5D_ |
|
not modelled |
20.8 |
10 |
PDB header:protein binding
Chain: D: PDB Molecule:pii protein;
PDBTitle: structural basis for the regulation of n-acetylglutamate kinase by pii2 in arabidopsis thaliana
|
| 120 | d2qwva1 |
|
not modelled |
20.7 |
17 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:AF1056-like |