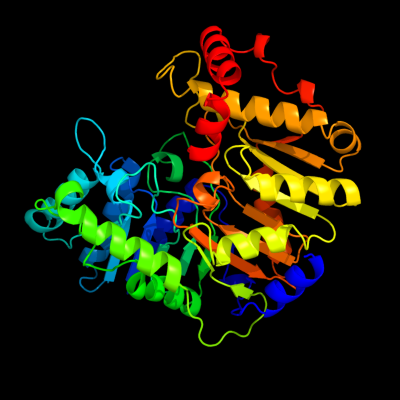
| 1 | c3hsiC_
|
|
 |
100.0 |
46 |
PDB header:transferase
Chain: C: PDB Molecule:phosphatidylserine synthase;
PDBTitle: crystal structure of phosphatidylserine synthase haemophilus2 influenzae rd kw20
|
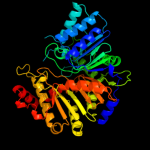
| 2 | c1v0sA_
|
|
 |
100.0 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:phospholipase d;
PDBTitle: uninhibited form of phospholipase d from streptomyces sp.2 strain pmf
|
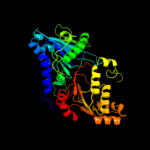
| 3 | d1v0wa1
|
|
 |
99.9 |
14 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Phospholipase D |
| 4 | c1xdoB_
|
|
 |
99.9 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:polyphosphate kinase;
PDBTitle: crystal structure of escherichia coli polyphosphate kinase
|
| 5 | d1xdpa3
|
|
 |
99.9 |
18 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Polyphosphate kinase C-terminal domain |
| 6 | d1byra_
|
|
 |
99.9 |
15 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Nuclease |
| 7 | d1v0wa2
|
|
 |
99.9 |
19 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Phospholipase D |
| 8 | c2o8rA_
|
|
 |
99.6 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:polyphosphate kinase;
PDBTitle: crystal structure of polyphosphate kinase from2 porphyromonas gingivalis
|
| 9 | d2o8ra3
|
|
 |
98.5 |
14 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Polyphosphate kinase C-terminal domain |
| 10 | d1xdpa4
|
|
 |
98.1 |
15 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Polyphosphate kinase C-terminal domain |
| 11 | c1q32C_
|
|
 |
97.9 |
14 |
PDB header:replication,transcription,hydrolase
Chain: C: PDB Molecule:tyrosyl-dna phosphodiesterase;
PDBTitle: crystal structure analysis of the yeast tyrosyl-dna2 phosphodiesterase
|
| 12 | c3sq3C_
|
|
 |
97.7 |
14 |
PDB header:hydrolase
Chain: C: PDB Molecule:tyrosyl-dna phosphodiesterase 1;
PDBTitle: crystal structure analysis of the yeast tyrosyl-dna phosphodiesterase2 h182a mutant
|
| 13 | d2o8ra4
|
|
 |
97.4 |
13 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Polyphosphate kinase C-terminal domain |
| 14 | c2c1lA_
|
|
 |
97.0 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:restriction endonuclease;
PDBTitle: structure of the bfii restriction endonuclease
|
| 15 | d1jy1a2
|
|
 |
95.8 |
30 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Tyrosyl-DNA phosphodiesterase TDP1 |
| 16 | c1nopB_
|
|
 |
95.6 |
35 |
PDB header:hydrolase/dna
Chain: B: PDB Molecule:tyrosyl-dna phosphodiesterase 1;
PDBTitle: crystal structure of human tyrosyl-dna phosphodiesterase2 (tdp1) in complex with vanadate, dna and a human3 topoisomerase i-derived peptide
|
| 17 | d1q32a2
|
|
 |
94.1 |
30 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Tyrosyl-DNA phosphodiesterase TDP1 |
| 18 | d2f5tx2
|
|
 |
62.9 |
16 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:TrmB middle domain-like |
| 19 | d1qzqa1
|
|
 |
62.8 |
29 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Tyrosyl-DNA phosphodiesterase TDP1 |
| 20 | d1q32a1
|
|
 |
51.7 |
11 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Tyrosyl-DNA phosphodiesterase TDP1 |
| 21 | c2f5tX_ |
|
not modelled |
51.3 |
16 |
PDB header:transcription
Chain: X: PDB Molecule:archaeal transcriptional regulator trmb;
PDBTitle: crystal structure of the sugar binding domain of the archaeal2 transcriptional regulator trmb
|
| 22 | d1jy1a1 |
|
not modelled |
38.2 |
14 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Tyrosyl-DNA phosphodiesterase TDP1 |
| 23 | c2a5hC_ |
|
not modelled |
37.2 |
11 |
PDB header:isomerase
Chain: C: PDB Molecule:l-lysine 2,3-aminomutase;
PDBTitle: 2.1 angstrom x-ray crystal structure of lysine-2,3-aminomutase from2 clostridium subterminale sb4, with michaelis analog (l-alpha-lysine3 external aldimine form of pyridoxal-5'-phosphate).
|
| 24 | d1vdda_ |
|
not modelled |
22.7 |
22 |
Fold:Recombination protein RecR
Superfamily:Recombination protein RecR
Family:Recombination protein RecR |
| 25 | c1vddC_ |
|
not modelled |
20.0 |
22 |
PDB header:recombination
Chain: C: PDB Molecule:recombination protein recr;
PDBTitle: crystal structure of recombinational repair protein recr
|
| 26 | d1f0ya2 |
|
not modelled |
18.8 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 27 | d1gz0a2 |
|
not modelled |
17.3 |
11 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:RNA 2'-O ribose methyltransferase substrate binding domain |
| 28 | c2x7mA_ |
|
not modelled |
17.3 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:archaemetzincin;
PDBTitle: crystal structure of archaemetzincin (amza) from methanopyrus2 kandleri at 1.5 a resolution
|
| 29 | d1f8ya_ |
|
not modelled |
16.8 |
6 |
Fold:Flavodoxin-like
Superfamily:N-(deoxy)ribosyltransferase-like
Family:N-deoxyribosyltransferase |
| 30 | c1t3gB_ |
|
not modelled |
16.7 |
8 |
PDB header:membrane protein
Chain: B: PDB Molecule:x-linked interleukin-1 receptor accessory
PDBTitle: crystal structure of the toll/interleukin-1 receptor (tir)2 domain of human il-1rapl
|
| 31 | c3hu5B_ |
|
not modelled |
16.5 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:isochorismatase family protein;
PDBTitle: crystal structure of isochorismatase family protein from desulfovibrio2 vulgaris subsp. vulgaris str. hildenborough
|
| 32 | d2bdea1 |
|
not modelled |
16.0 |
24 |
Fold:HAD-like
Superfamily:HAD-like
Family:5' nucleotidase-like |
| 33 | d1s2da_ |
|
not modelled |
16.0 |
14 |
Fold:Flavodoxin-like
Superfamily:N-(deoxy)ribosyltransferase-like
Family:N-deoxyribosyltransferase |
| 34 | d1e8ob_ |
|
not modelled |
15.8 |
11 |
Fold:Signal recognition particle alu RNA binding heterodimer, SRP9/14
Superfamily:Signal recognition particle alu RNA binding heterodimer, SRP9/14
Family:Signal recognition particle alu RNA binding heterodimer, SRP9/14 |
| 35 | c1b9xC_ |
|
not modelled |
15.5 |
21 |
PDB header:signaling protein
Chain: C: PDB Molecule:protein (phosducin);
PDBTitle: structural analysis of phosducin and its phosphorylation-2 regulated interaction with transducin
|
| 36 | c2l82A_ |
|
not modelled |
15.5 |
26 |
PDB header:de novo protein
Chain: A: PDB Molecule:designed protein or32;
PDBTitle: solution nmr structure of de novo designed protein, p-loop ntpase2 fold, northeast structural genomics consortium target or32
|
| 37 | d1x9ga_ |
|
not modelled |
13.9 |
14 |
Fold:Isochorismatase-like hydrolases
Superfamily:Isochorismatase-like hydrolases
Family:Isochorismatase-like hydrolases |
| 38 | d1gz0f2 |
|
not modelled |
13.0 |
11 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:RNA 2'-O ribose methyltransferase substrate binding domain |
| 39 | d1914a2 |
|
not modelled |
11.5 |
12 |
Fold:Signal recognition particle alu RNA binding heterodimer, SRP9/14
Superfamily:Signal recognition particle alu RNA binding heterodimer, SRP9/14
Family:Signal recognition particle alu RNA binding heterodimer, SRP9/14 |
| 40 | d1j5pa4 |
|
not modelled |
11.4 |
9 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 41 | c1zrwA_ |
|
not modelled |
10.9 |
46 |
PDB header:antimicrobial protein, antibiotic
Chain: A: PDB Molecule:spinigerin;
PDBTitle: solution structure of spinigerin in h20/tfe 10%
|
| 42 | c1zrvA_ |
|
not modelled |
10.9 |
46 |
PDB header:antimicrobial protein, antibiotic
Chain: A: PDB Molecule:spinigerin;
PDBTitle: solution structure of spinigerin in h20/tfe 50%
|
| 43 | c2b34C_ |
|
not modelled |
10.4 |
16 |
PDB header:hydrolase
Chain: C: PDB Molecule:mar1 ribonuclease;
PDBTitle: structure of mar1 ribonuclease from caenorhabditis elegans
|
| 44 | c2jcmA_ |
|
not modelled |
10.0 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:cytosolic purine 5'-nucleotidase;
PDBTitle: crystal structure of human cytosolic 5'-nucleotidase ii in2 complex with beryllium trifluoride
|
| 45 | c3gbcA_ |
|
not modelled |
9.9 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:pyrazinamidase/nicotinamidas pnca;
PDBTitle: determination of the crystal structure of the pyrazinamidase from2 m.tuberculosis : a structure-function analysis for prediction3 resistance to pyrazinamide
|
| 46 | c2r60A_ |
|
not modelled |
9.4 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:glycosyl transferase, group 1;
PDBTitle: structure of apo sucrose phosphate synthase (sps) of2 halothermothrix orenii
|
| 47 | c2jz2A_ |
|
not modelled |
9.4 |
23 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:ssl0352 protein;
PDBTitle: solution nmr structure of ssl0352 protein from synechocystis sp. pcc2 6803. northeast structural genomics consortium target sgr42
|
| 48 | d1ldna1 |
|
not modelled |
9.3 |
9 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 49 | c3l2iB_ |
|
not modelled |
9.3 |
8 |
PDB header:lyase
Chain: B: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: 1.85 angstrom crystal structure of the 3-dehydroquinate dehydratase2 (arod) from salmonella typhimurium lt2.
|
| 50 | c2kxeA_ |
|
not modelled |
9.2 |
29 |
PDB header:transferase
Chain: A: PDB Molecule:dna polymerase ii small subunit;
PDBTitle: n-terminal domain of the dp1 subunit of an archaeal d-family dna2 polymerase
|
| 51 | c1g4wR_ |
|
not modelled |
8.9 |
11 |
PDB header:signaling protein
Chain: R: PDB Molecule:protein tyrosine phosphatase sptp;
PDBTitle: crystal structure of the salmonella tyrosine phosphatase2 and gtpase activating protein sptp
|
| 52 | d1m6ia2 |
|
not modelled |
8.8 |
19 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 53 | d1guza1 |
|
not modelled |
8.5 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 54 | c3ua4A_ |
|
not modelled |
8.1 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:protein arginine n-methyltransferase 5;
PDBTitle: crystal structure of protein arginine methyltransferase prmt5
|
| 55 | c3dy0B_ |
|
not modelled |
7.9 |
20 |
PDB header:blood clotting, hydrolase inhibitor
Chain: B: PDB Molecule:c-terminus plasma serine protease inhibitor;
PDBTitle: crystal structure of cleaved pci bound to heparin
|
| 56 | c1xeaD_ |
|
not modelled |
7.8 |
8 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of a gfo/idh/moca family oxidoreductase2 from vibrio cholerae
|
| 57 | d1ojua1 |
|
not modelled |
7.7 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 58 | d1im5a_ |
|
not modelled |
7.5 |
11 |
Fold:Isochorismatase-like hydrolases
Superfamily:Isochorismatase-like hydrolases
Family:Isochorismatase-like hydrolases |
| 59 | c3gt0A_ |
|
not modelled |
7.4 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pyrroline-5-carboxylate reductase;
PDBTitle: crystal structure of pyrroline 5-carboxylate reductase from bacillus2 cereus. northeast structural genomics consortium target bcr38b
|
| 60 | c1lq8H_ |
|
not modelled |
7.4 |
20 |
PDB header:blood clotting
Chain: H: PDB Molecule:plasma serine protease inhibitor;
PDBTitle: crystal structure of cleaved protein c inhibitor
|
| 61 | d2g8la1 |
|
not modelled |
7.2 |
24 |
Fold:AF1104-like
Superfamily:AF1104-like
Family:AF1104-like |
| 62 | c3oqhB_ |
|
not modelled |
7.0 |
16 |
PDB header:ligase
Chain: B: PDB Molecule:putative uncharacterized protein yvmc;
PDBTitle: crystal structure of b. licheniformis cdps yvmc-blic
|
| 63 | d1czan3 |
|
not modelled |
6.9 |
22 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Hexokinase |
| 64 | d5ldha1 |
|
not modelled |
6.7 |
9 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 65 | d1x94a_ |
|
not modelled |
6.4 |
10 |
Fold:SIS domain
Superfamily:SIS domain
Family:mono-SIS domain |
| 66 | d1sjpa3 |
|
not modelled |
6.3 |
6 |
Fold:GroEL-intermediate domain like
Superfamily:GroEL-intermediate domain like
Family:GroEL-like chaperone, intermediate domain |
| 67 | d1obba1 |
|
not modelled |
6.3 |
11 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 68 | d1llda1 |
|
not modelled |
6.3 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 69 | d1gqna_ |
|
not modelled |
6.3 |
6 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 70 | c3qufB_ |
|
not modelled |
6.3 |
14 |
PDB header:transport protein
Chain: B: PDB Molecule:extracellular solute-binding protein, family 1;
PDBTitle: the structure of a family 1 extracellular solute-binding protein from2 bifidobacterium longum subsp. infantis
|
| 71 | d1ez4a1 |
|
not modelled |
6.2 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 72 | d1u8xx1 |
|
not modelled |
6.2 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 73 | d1up7a1 |
|
not modelled |
6.1 |
10 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 74 | d2zdra2 |
|
not modelled |
6.1 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:NeuB-like |
| 75 | c1llcA_ |
|
not modelled |
6.1 |
14 |
PDB header:oxidoreductase(choh(d)-nad(a))
Chain: A: PDB Molecule:l-lactate dehydrogenase;
PDBTitle: structure determination of the allosteric l-lactate dehydrogenase from2 lactobacillus casei at 3.0 angstroms resolution
|
| 76 | c2x0dA_ |
|
not modelled |
6.1 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:wsaf;
PDBTitle: apo structure of wsaf
|
| 77 | d1jjcb4 |
|
not modelled |
6.1 |
13 |
Fold:Ferredoxin-like
Superfamily:Anticodon-binding domain of PheRS
Family:Anticodon-binding domain of PheRS |
| 78 | d1a0rp_ |
|
not modelled |
6.1 |
21 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Phosducin |
| 79 | c2v6bB_ |
|
not modelled |
5.7 |
7 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:l-lactate dehydrogenase;
PDBTitle: crystal structure of lactate dehydrogenase from deinococcus2 radiodurans (apo form)
|
| 80 | c2yvaB_ |
|
not modelled |
5.7 |
7 |
PDB header:dna binding protein
Chain: B: PDB Molecule:dnaa initiator-associating protein diaa;
PDBTitle: crystal structure of escherichia coli diaa
|
| 81 | c2qupA_ |
|
not modelled |
5.7 |
9 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:bh1478 protein;
PDBTitle: crystal structure of uncharacterized protein bh1478 from bacillus2 halodurans
|
| 82 | c2krcA_ |
|
not modelled |
5.6 |
17 |
PDB header:transcription
Chain: A: PDB Molecule:dna-directed rna polymerase subunit delta;
PDBTitle: solution structure of the n-terminal domain of bacillus2 subtilis delta subunit of rna polymerase
|
| 83 | d4pgaa_ |
|
not modelled |
5.6 |
12 |
Fold:Glutaminase/Asparaginase
Superfamily:Glutaminase/Asparaginase
Family:Glutaminase/Asparaginase |
| 84 | c1a5zA_ |
|
not modelled |
5.6 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-lactate dehydrogenase;
PDBTitle: lactate dehydrogenase from thermotoga maritima (tmldh)
|
| 85 | d1llca1 |
|
not modelled |
5.6 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 86 | d1ldma1 |
|
not modelled |
5.5 |
12 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 87 | c3js3C_ |
|
not modelled |
5.5 |
8 |
PDB header:lyase
Chain: C: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: crystal structure of type i 3-dehydroquinate dehydratase (arod) from2 clostridium difficile with covalent reaction intermediate
|
| 88 | d2dx5a1 |
|
not modelled |
5.5 |
19 |
Fold:PH domain-like barrel
Superfamily:PH domain-like
Family:VPS36 N-terminal domain-like |
| 89 | c3bsmD_ |
|
not modelled |
5.5 |
12 |
PDB header:lyase
Chain: D: PDB Molecule:mandelate racemase/muconate lactonizing enzyme;
PDBTitle: crystal structure of d-mannonate dehydratase from2 chromohalobacter salexigens
|
| 90 | d1x92a_ |
|
not modelled |
5.4 |
15 |
Fold:SIS domain
Superfamily:SIS domain
Family:mono-SIS domain |
| 91 | d2bisa1 |
|
not modelled |
5.4 |
15 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:Glycosyl transferases group 1 |
| 92 | c1914A_ |
|
not modelled |
5.3 |
12 |
PDB header:alu domain
Chain: A: PDB Molecule:signal recognition particle 9/14 fusion protein;
PDBTitle: signal recognition particle alu rna binding heterodimer, srp9/14
|
| 93 | d2p61a1 |
|
not modelled |
5.3 |
7 |
Fold:Four-helical up-and-down bundle
Superfamily:TM1646-like
Family:TM1646-like |
| 94 | c2pr7A_ |
|
not modelled |
5.3 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:haloacid dehalogenase/epoxide hydrolase family;
PDBTitle: crystal structure of uncharacterized protein (np_599989.1) from2 corynebacterium glutamicum atcc 13032 kitasato at 1.44 a resolution
|
| 95 | d2bf5a1 |
|
not modelled |
5.2 |
14 |
Fold:Monooxygenase (hydroxylase) regulatory protein
Superfamily:Monooxygenase (hydroxylase) regulatory protein
Family:Monooxygenase (hydroxylase) regulatory protein |
| 96 | c2x3yA_ |
|
not modelled |
5.2 |
7 |
PDB header:isomerase
Chain: A: PDB Molecule:phosphoheptose isomerase;
PDBTitle: crystal structure of gmha from burkholderia pseudomallei
|
| 97 | c2aj6A_ |
|
not modelled |
5.1 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:hypothetical protein mw0638;
PDBTitle: crystal structure of a putative gnat family acetyltransferase (mw0638)2 from staphylococcus aureus subsp. aureus at 1.63 a resolution
|
| 98 | c2p61A_ |
|
not modelled |
5.1 |
7 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein tm_1646;
PDBTitle: crystal structure of protein tm1646 from thermotoga2 maritima, pfam duf327
|