| 1 | c2vlbC_
|
|
 |
99.4 |
19 |
PDB header:lyase
Chain: C: PDB Molecule:arylmalonate decarboxylase;
PDBTitle: structure of unliganded arylmalonate decarboxylase
|
| 2 | c3qvjB_
|
|
 |
98.8 |
13 |
PDB header:isomerase
Chain: B: PDB Molecule:putative hydantoin racemase;
PDBTitle: allantoin racemase from klebsiella pneumoniae
|
| 3 | c2dgdD_
|
|
 |
98.8 |
16 |
PDB header:lyase
Chain: D: PDB Molecule:223aa long hypothetical arylmalonate decarboxylase;
PDBTitle: crystal structure of st0656, a function unknown protein from2 sulfolobus tokodaii
|
| 4 | c2eq5D_
|
|
 |
98.6 |
18 |
PDB header:isomerase
Chain: D: PDB Molecule:228aa long hypothetical hydantoin racemase;
PDBTitle: crystal structure of hydantoin racemase from pyrococcus horikoshii ot3
|
| 5 | c2xecD_
|
|
 |
98.5 |
15 |
PDB header:isomerase
Chain: D: PDB Molecule:putative maleate isomerase;
PDBTitle: nocardia farcinica maleate cis-trans isomerase bound to2 tris
|
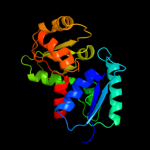
| 6 | c2gzmB_
|
|
 |
97.9 |
17 |
PDB header:isomerase
Chain: B: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of the glutamate racemase from bacillus2 anthracis
|
| 7 | c3outC_
|
|
 |
97.9 |
16 |
PDB header:isomerase
Chain: C: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of glutamate racemase from francisella tularensis2 subsp. tularensis schu s4 in complex with d-glutamate.
|
| 8 | c2dwuA_
|
|
 |
97.8 |
19 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of glutamate racemase isoform race1 from bacillus2 anthracis
|
| 9 | c1b74A_
|
|
 |
97.8 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: glutamate racemase from aquifex pyrophilus
|
| 10 | c2jfoB_
|
|
 |
97.7 |
13 |
PDB header:isomerase
Chain: B: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of enterococcus faecalis glutamate2 racemase in complex with d- and l-glutamate
|
| 11 | c3s81A_
|
|
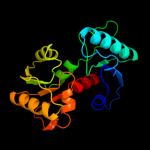 |
97.7 |
16 |
PDB header:isomerase
Chain: A: PDB Molecule:putative aspartate racemase;
PDBTitle: crystal structure of putative aspartate racemase from salmonella2 typhimurium
|
| 12 | c3hfrA_
|
|
 |
97.5 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of glutamate racemase from listeria monocytogenes
|
| 13 | c2jfqA_
|
|
 |
97.5 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of staphylococcus aureus glutamate2 racemase in complex with d-glutamate
|
| 14 | c1zuwA_
|
|
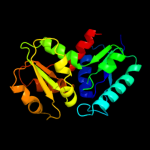 |
97.3 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase 1;
PDBTitle: crystal structure of b.subtilis glutamate racemase (race) with d-glu
|
| 15 | c2ohoA_
|
|
 |
97.2 |
20 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: structural basis for glutamate racemase inhibitor
|
| 16 | c2jfzB_
|
|
 |
96.8 |
11 |
PDB header:isomerase
Chain: B: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of helicobacter pylori glutamate racemase2 in complex with d-glutamate and an inhibitor
|
| 17 | c3ojcD_
|
|
 |
96.6 |
20 |
PDB header:isomerase
Chain: D: PDB Molecule:putative aspartate/glutamate racemase;
PDBTitle: crystal structure of a putative asp/glu racemase from yersinia pestis
|
| 18 | c2dx7B_
|
|
 |
96.4 |
14 |
PDB header:isomerase
Chain: B: PDB Molecule:aspartate racemase;
PDBTitle: crystal structure of pyrococcus horikoshii ot3 aspartate racemase2 complex with citric acid
|
| 19 | c2zskA_
|
|
 |
96.1 |
15 |
PDB header:unknown function
Chain: A: PDB Molecule:226aa long hypothetical aspartate racemase;
PDBTitle: crystal structure of ph1733, an aspartate racemase2 homologue, from pyrococcus horikoshii ot3
|
| 20 | c2jfnA_
|
|
 |
95.5 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of escherichia coli glutamate racemase2 in complex with l-glutamate and activator udp-murnac-ala
|
| 21 | c3bg3B_ |
|
not modelled |
93.2 |
15 |
PDB header:ligase
Chain: B: PDB Molecule:pyruvate carboxylase, mitochondrial;
PDBTitle: crystal structure of human pyruvate carboxylase (missing2 the biotin carboxylase domain at the n-terminus)
|
| 22 | c3ez4B_ |
|
not modelled |
91.3 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:3-methyl-2-oxobutanoate hydroxymethyltransferase;
PDBTitle: crystal structure of 3-methyl-2-oxobutanoate2 hydroxymethyltransferase from burkholderia pseudomallei
|
| 23 | d1oy0a_ |
|
not modelled |
90.4 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Ketopantoate hydroxymethyltransferase PanB |
| 24 | d1m3ua_ |
|
not modelled |
90.4 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Ketopantoate hydroxymethyltransferase PanB |
| 25 | c1nvmG_ |
|
not modelled |
88.6 |
18 |
PDB header:lyase/oxidoreductase
Chain: G: PDB Molecule:4-hydroxy-2-oxovalerate aldolase;
PDBTitle: crystal structure of a bifunctional aldolase-dehydrogenase :2 sequestering a reactive and volatile intermediate
|
| 26 | d1jfla1 |
|
not modelled |
88.3 |
26 |
Fold:ATC-like
Superfamily:Aspartate/glutamate racemase
Family:Aspartate/glutamate racemase |
| 27 | d1nvma2 |
|
not modelled |
88.0 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:HMGL-like |
| 28 | c2hqbA_ |
|
not modelled |
86.8 |
20 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional activator of comk gene;
PDBTitle: crystal structure of a transcriptional activator of comk2 gene from bacillus halodurans
|
| 29 | c3bleA_ |
|
not modelled |
86.1 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:citramalate synthase from leptospira interrogans;
PDBTitle: crystal structure of the catalytic domain of licms in2 complexed with malonate
|
| 30 | d1jfla2 |
|
not modelled |
84.0 |
20 |
Fold:ATC-like
Superfamily:Aspartate/glutamate racemase
Family:Aspartate/glutamate racemase |
| 31 | d1muma_ |
|
not modelled |
83.6 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 32 | d2dx7a1 |
|
not modelled |
81.6 |
19 |
Fold:ATC-like
Superfamily:Aspartate/glutamate racemase
Family:Aspartate/glutamate racemase |
| 33 | d1j8yf2 |
|
not modelled |
81.3 |
15 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
| 34 | d1sr9a2 |
|
not modelled |
80.3 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:HMGL-like |
| 35 | d2ajta2 |
|
not modelled |
78.4 |
11 |
Fold:FucI/AraA N-terminal and middle domains
Superfamily:FucI/AraA N-terminal and middle domains
Family:AraA N-terminal and middle domain-like |
| 36 | c3s5oA_ |
|
not modelled |
77.5 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:4-hydroxy-2-oxoglutarate aldolase, mitochondrial;
PDBTitle: crystal structure of human 4-hydroxy-2-oxoglutarate aldolase bound to2 pyruvate
|
| 37 | c3a23A_ |
|
not modelled |
74.4 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative secreted alpha-galactosidase;
PDBTitle: crystal structure of beta-l-arabinopyranosidase complexed with d-2 galactose
|
| 38 | c2og2A_ |
|
not modelled |
71.2 |
16 |
PDB header:protein transport
Chain: A: PDB Molecule:putative signal recognition particle receptor;
PDBTitle: crystal structure of chloroplast ftsy from arabidopsis2 thaliana
|
| 39 | c2ftpA_ |
|
not modelled |
70.2 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:hydroxymethylglutaryl-coa lyase;
PDBTitle: crystal structure of hydroxymethylglutaryl-coa lyase from pseudomonas2 aeruginosa
|
| 40 | c3dhyC_ |
|
not modelled |
69.4 |
22 |
PDB header:hydrolase
Chain: C: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structures of mycobacterium tuberculosis s-adenosyl-l-2 homocysteine hydrolase in ternary complex with substrate and3 inhibitors
|
| 41 | c2wdfA_ |
|
not modelled |
69.3 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:sulfur oxidation protein soxb;
PDBTitle: termus thermophilus sulfate thiohydrolase soxb
|
| 42 | d2p10a1 |
|
not modelled |
68.9 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Mll9387-like |
| 43 | d1tqja_ |
|
not modelled |
68.9 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
| 44 | c1ydnA_ |
|
not modelled |
66.3 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:hydroxymethylglutaryl-coa lyase;
PDBTitle: crystal structure of the hmg-coa lyase from brucella melitensis,2 northeast structural genomics target lr35.
|
| 45 | c2vc6A_ |
|
not modelled |
65.2 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: structure of mosa from s. meliloti with pyruvate bound
|
| 46 | d1b74a2 |
|
not modelled |
64.9 |
11 |
Fold:ATC-like
Superfamily:Aspartate/glutamate racemase
Family:Aspartate/glutamate racemase |
| 47 | c3h5dD_ |
|
not modelled |
64.8 |
15 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: dihydrodipicolinate synthase from drug-resistant streptococcus2 pneumoniae
|
| 48 | c3zu0A_ |
|
not modelled |
64.2 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:nad nucleotidase;
PDBTitle: structure of haemophilus influenzae nad nucleotidase (nadn)
|
| 49 | c3navB_ |
|
not modelled |
64.0 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of an alpha subunit of tryptophan synthase from2 vibrio cholerae o1 biovar el tor str. n16961
|
| 50 | c2z1aA_ |
|
not modelled |
63.4 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:5'-nucleotidase;
PDBTitle: crystal structure of 5'-nucleotidase precursor from thermus2 thermophilus hb8
|
| 51 | c3eooL_ |
|
not modelled |
63.0 |
18 |
PDB header:lyase
Chain: L: PDB Molecule:methylisocitrate lyase;
PDBTitle: 2.9a crystal structure of methyl-isocitrate lyase from2 burkholderia pseudomallei
|
| 52 | c3dmdA_ |
|
not modelled |
62.6 |
15 |
PDB header:transport protein
Chain: A: PDB Molecule:signal recognition particle receptor;
PDBTitle: structures and conformations in solution of the signal recognition2 particle receptor from the archaeon pyrococcus furiosus
|
| 53 | d1o66a_ |
|
not modelled |
61.4 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Ketopantoate hydroxymethyltransferase PanB |
| 54 | c3dz1A_ |
|
not modelled |
61.1 |
11 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 1.87a resolution
|
| 55 | c3e96B_ |
|
not modelled |
60.1 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 bacillus clausii
|
| 56 | c3ivuB_ |
|
not modelled |
60.0 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:homocitrate synthase, mitochondrial;
PDBTitle: homocitrate synthase lys4 bound to 2-og
|
| 57 | c2p10D_ |
|
not modelled |
58.8 |
13 |
PDB header:hydrolase
Chain: D: PDB Molecule:mll9387 protein;
PDBTitle: crystal structure of a putative phosphonopyruvate hydrolase (mll9387)2 from mesorhizobium loti maff303099 at 2.15 a resolution
|
| 58 | c1zlpA_ |
|
not modelled |
58.6 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:petal death protein;
PDBTitle: petal death protein psr132 with cysteine-linked glutaraldehyde forming2 a thiohemiacetal adduct
|
| 59 | c1zfjA_ |
|
not modelled |
57.8 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine monophosphate dehydrogenase;
PDBTitle: inosine monophosphate dehydrogenase (impdh; ec 1.1.1.205) from2 streptococcus pyogenes
|
| 60 | c3o1hB_ |
|
not modelled |
56.3 |
13 |
PDB header:signaling protein
Chain: B: PDB Molecule:periplasmic protein tort;
PDBTitle: crystal structure of the tors sensor domain - tort complex in the2 presence of tmao
|
| 61 | c2cnwF_ |
|
not modelled |
56.0 |
19 |
PDB header:signal recognition
Chain: F: PDB Molecule:cell division protein ftsy;
PDBTitle: gdpalf4 complex of the srp gtpases ffh and ftsy
|
| 62 | d1g2oa_ |
|
not modelled |
55.5 |
16 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 63 | c2vsnB_ |
|
not modelled |
54.9 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:xcogt;
PDBTitle: structure and topological arrangement of an o-glcnac2 transferase homolog: insight into molecular control of3 intracellular glycosylation
|
| 64 | c2j7pA_ |
|
not modelled |
54.5 |
16 |
PDB header:signal recognition
Chain: A: PDB Molecule:signal recognition particle protein;
PDBTitle: gmppnp-stabilized ng domain complex of the srp gtpases ffh2 and ftsy
|
| 65 | c3hcwB_ |
|
not modelled |
54.4 |
13 |
PDB header:rna binding protein
Chain: B: PDB Molecule:maltose operon transcriptional repressor;
PDBTitle: crystal structure of probable maltose operon transcriptional repressor2 malr from staphylococcus areus
|
| 66 | c3bg5C_ |
|
not modelled |
53.9 |
11 |
PDB header:ligase
Chain: C: PDB Molecule:pyruvate carboxylase;
PDBTitle: crystal structure of staphylococcus aureus pyruvate2 carboxylase
|
| 67 | d1chma1 |
|
not modelled |
53.7 |
8 |
Fold:Ribonuclease H-like motif
Superfamily:Creatinase/prolidase N-terminal domain
Family:Creatinase/prolidase N-terminal domain |
| 68 | c3lrmB_ |
|
not modelled |
52.7 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:alpha-galactosidase 1;
PDBTitle: structure of alfa-galactosidase from saccharomyces cerevisiae with2 raffinose
|
| 69 | c2qygC_ |
|
not modelled |
52.6 |
16 |
PDB header:unknown function
Chain: C: PDB Molecule:ribulose bisphosphate carboxylase-like protein 2;
PDBTitle: crystal structure of a rubisco-like protein rlp2 from rhodopseudomonas2 palustris
|
| 70 | c1yb2A_ |
|
not modelled |
51.8 |
10 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein ta0852;
PDBTitle: structure of a putative methyltransferase from thermoplasma2 acidophilum.
|
| 71 | d1yb2a1 |
|
not modelled |
51.8 |
10 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:tRNA(1-methyladenosine) methyltransferase-like |
| 72 | c3daqB_ |
|
not modelled |
51.3 |
25 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from methicillin-2 resistant staphylococcus aureus
|
| 73 | d1ykwa1 |
|
not modelled |
50.9 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
| 74 | c3gndC_ |
|
not modelled |
50.5 |
14 |
PDB header:lyase
Chain: C: PDB Molecule:aldolase lsrf;
PDBTitle: crystal structure of e. coli lsrf in complex with ribulose-5-phosphate
|
| 75 | c3lyeA_ |
|
not modelled |
50.1 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:oxaloacetate acetyl hydrolase;
PDBTitle: crystal structure of oxaloacetate acetylhydrolase
|
| 76 | c3lciA_ |
|
not modelled |
49.8 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:n-acetylneuraminate lyase;
PDBTitle: the d-sialic acid aldolase mutant v251w
|
| 77 | d1li4a2 |
|
not modelled |
48.1 |
13 |
Fold:Flavodoxin-like
Superfamily:Formate/glycerate dehydrogenase catalytic domain-like
Family:S-adenosylhomocystein hydrolase |
| 78 | c3pe3D_ |
|
not modelled |
47.9 |
12 |
PDB header:transferase
Chain: D: PDB Molecule:udp-n-acetylglucosamine--peptide n-
PDBTitle: structure of human o-glcnac transferase and its complex with a peptide2 substrate
|
| 79 | d1o4va_ |
|
not modelled |
47.5 |
20 |
Fold:Flavodoxin-like
Superfamily:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)
Family:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE) |
| 80 | c3n58D_ |
|
not modelled |
47.4 |
18 |
PDB header:hydrolase
Chain: D: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structure of s-adenosyl-l-homocysteine hydrolase from brucella2 melitensis in ternary complex with nad and adenosine, orthorhombic3 form
|
| 81 | c1xuzA_ |
|
not modelled |
47.2 |
19 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:polysialic acid capsule biosynthesis protein siac;
PDBTitle: crystal structure analysis of sialic acid synthase (neub)from2 neisseria meningitidis, bound to mn2+, phosphoenolpyruvate, and n-3 acetyl mannosaminitol
|
| 82 | d1fuia2 |
|
not modelled |
47.0 |
3 |
Fold:FucI/AraA N-terminal and middle domains
Superfamily:FucI/AraA N-terminal and middle domains
Family:L-fucose isomerase, N-terminal and second domains |
| 83 | c3s99A_ |
|
not modelled |
46.9 |
14 |
PDB header:lipid binding protein
Chain: A: PDB Molecule:basic membrane lipoprotein;
PDBTitle: crystal structure of a basic membrane lipoprotein from brucella2 melitensis, iodide soak
|
| 84 | d1dbqa_ |
|
not modelled |
45.9 |
11 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
| 85 | d2zdra2 |
|
not modelled |
45.5 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:NeuB-like |
| 86 | d1meoa_ |
|
not modelled |
44.6 |
9 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
| 87 | d1b74a1 |
|
not modelled |
44.5 |
22 |
Fold:ATC-like
Superfamily:Aspartate/glutamate racemase
Family:Aspartate/glutamate racemase |
| 88 | c2qiwA_ |
|
not modelled |
44.3 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:pep phosphonomutase;
PDBTitle: crystal structure of a putative phosphoenolpyruvate phosphonomutase2 (ncgl1015, cgl1060) from corynebacterium glutamicum atcc 13032 at3 1.80 a resolution
|
| 89 | c3gv0A_ |
|
not modelled |
43.8 |
11 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator, laci family;
PDBTitle: crystal structure of laci family transcription regulator from2 agrobacterium tumefaciens
|
| 90 | c3fluD_ |
|
not modelled |
43.3 |
13 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from the pathogen2 neisseria meningitidis
|
| 91 | c2ehhE_ |
|
not modelled |
43.1 |
10 |
PDB header:lyase
Chain: E: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 aquifex aeolicus
|
| 92 | c3raoB_ |
|
not modelled |
43.1 |
11 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative luciferase-like monooxygenase;
PDBTitle: crystal structure of the luciferase-like monooxygenase from bacillus2 cereus atcc 10987.
|
| 93 | c3fkkA_ |
|
not modelled |
42.5 |
11 |
PDB header:lyase
Chain: A: PDB Molecule:l-2-keto-3-deoxyarabonate dehydratase;
PDBTitle: structure of l-2-keto-3-deoxyarabonate dehydratase
|
| 94 | c2qmoA_ |
|
not modelled |
41.5 |
13 |
PDB header:ligase
Chain: A: PDB Molecule:dethiobiotin synthetase;
PDBTitle: crystal structure of dethiobiotin synthetase (biod) from helicobacter2 pylori
|
| 95 | c3g8rA_ |
|
not modelled |
41.1 |
19 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:probable spore coat polysaccharide biosynthesis protein e;
PDBTitle: crystal structure of putative spore coat polysaccharide biosynthesis2 protein e from chromobacterium violaceum atcc 12472
|
| 96 | c1telA_ |
|
not modelled |
41.0 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:ribulose bisphosphate carboxylase, large subunit;
PDBTitle: crystal structure of a rubisco-like protein from chlorobium2 tepidum
|
| 97 | c2yv4A_ |
|
not modelled |
40.9 |
14 |
PDB header:rna binding protein
Chain: A: PDB Molecule:hypothetical protein ph0435;
PDBTitle: crystal structure of c-terminal sua5 domain from pyrococcus horikoshii2 hypothetical sua5 protein ph0435
|
| 98 | d2z1aa2 |
|
not modelled |
40.8 |
24 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:5'-nucleotidase (syn. UDP-sugar hydrolase), N-terminal domain |
| 99 | d1tlfa_ |
|
not modelled |
40.5 |
18 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
| 100 | c3eegB_ |
|
not modelled |
40.3 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:2-isopropylmalate synthase;
PDBTitle: crystal structure of a 2-isopropylmalate synthase from2 cytophaga hutchinsonii
|
| 101 | d1ps9a1 |
|
not modelled |
39.8 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 102 | d1hl2a_ |
|
not modelled |
37.8 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 103 | c3h14A_ |
|
not modelled |
37.6 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:aminotransferase, classes i and ii;
PDBTitle: crystal structure of a putative aminotransferase from silicibacter2 pomeroyi
|
| 104 | d1h1ya_ |
|
not modelled |
36.7 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
| 105 | c3fa4D_ |
|
not modelled |
36.4 |
23 |
PDB header:lyase
Chain: D: PDB Molecule:2,3-dimethylmalate lyase;
PDBTitle: crystal structure of 2,3-dimethylmalate lyase, a pep mutase/isocitrate2 lyase superfamily member, triclinic crystal form
|
| 106 | c3si9B_ |
|
not modelled |
36.2 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from bartonella2 henselae
|
| 107 | c3gveB_ |
|
not modelled |
36.1 |
27 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:yfkn protein;
PDBTitle: crystal structure of calcineurin-like phosphoesterase yfkn from2 bacillus subtilis
|
| 108 | c3gkaB_ |
|
not modelled |
35.7 |
12 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:n-ethylmaleimide reductase;
PDBTitle: crystal structure of n-ethylmaleimidine reductase from2 burkholderia pseudomallei
|
| 109 | d1ujqa_ |
|
not modelled |
35.5 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 110 | c3b9qA_ |
|
not modelled |
35.2 |
18 |
PDB header:protein transport
Chain: A: PDB Molecule:chloroplast srp receptor homolog, alpha subunit
PDBTitle: the crystal structure of cpftsy from arabidopsis thaliana
|
| 111 | c2h31A_ |
|
not modelled |
35.0 |
11 |
PDB header:ligase, lyase
Chain: A: PDB Molecule:multifunctional protein ade2;
PDBTitle: crystal structure of human paics, a bifunctional carboxylase and2 synthetase in purine biosynthesis
|
| 112 | c3d64A_ |
|
not modelled |
34.8 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structure of s-adenosyl-l-homocysteine hydrolase from2 burkholderia pseudomallei
|
| 113 | c3eb2A_ |
|
not modelled |
34.7 |
11 |
PDB header:lyase
Chain: A: PDB Molecule:putative dihydrodipicolinate synthetase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 2.0a resolution
|
| 114 | d1nqka_ |
|
not modelled |
34.5 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Ssud-like monoxygenases |
| 115 | d1v8ba2 |
|
not modelled |
34.3 |
6 |
Fold:Flavodoxin-like
Superfamily:Formate/glycerate dehydrogenase catalytic domain-like
Family:S-adenosylhomocystein hydrolase |
| 116 | d1qcza_ |
|
not modelled |
34.0 |
12 |
Fold:Flavodoxin-like
Superfamily:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)
Family:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE) |
| 117 | c1j8yF_ |
|
not modelled |
33.2 |
18 |
PDB header:signaling protein
Chain: F: PDB Molecule:signal recognition 54 kda protein;
PDBTitle: signal recognition particle conserved gtpase domain from a.2 ambivalens t112a mutant
|
| 118 | c3d0cB_ |
|
not modelled |
33.1 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 oceanobacillus iheyensis at 1.9 a resolution
|
| 119 | c2rfgB_ |
|
not modelled |
32.4 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from hahella2 chejuensis at 1.5a resolution
|
| 120 | c3noeA_ |
|
not modelled |
32.1 |
8 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from pseudomonas2 aeruginosa
|