| 1 | c1hk8A_ |
|
 |
100.0 |
57 |
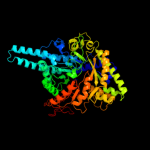
PDB header:oxidoreductase
Chain: A: PDB Molecule:anaerobic ribonucleotide-triphosphate reductase;
PDBTitle: structural basis for allosteric substrate specificity2 regulation in class iii ribonucleotide reductases:3 nrdd in complex with dgtp
|
| 2 | d1hk8a_ |
|
 |
100.0 |
57 |
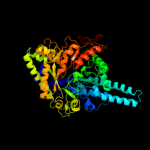
Fold:PFL-like glycyl radical enzymes
Superfamily:PFL-like glycyl radical enzymes
Family:Class III anaerobic ribonucleotide reductase NRDD subunit |
| 3 | c3r1rB_ |
|
 |
99.9 |
13 |
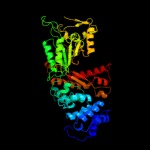
PDB header:complex (oxidoreductase/peptide)
Chain: B: PDB Molecule:ribonucleotide reductase r1 protein;
PDBTitle: ribonucleotide reductase r1 protein with amppnp occupying2 the activity site from escherichia coli
|
| 4 | c3hnfA_ |
|
 |
99.8 |
14 |
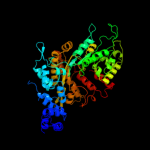
PDB header:oxidoreductase
Chain: A: PDB Molecule:ribonucleoside-diphosphate reductase large subunit;
PDBTitle: crystal structure of human ribonucleotide reductase 1 bound to the2 effectors ttp and datp
|
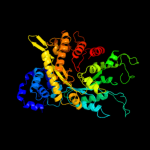
| 5 | c2cvuA_ |
|
 |
99.8 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ribonucleoside-diphosphate reductase large chain
PDBTitle: structures of yeast ribonucleotide reductase i
|
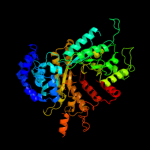
| 6 | c2wghA_ |
|
 |
99.8 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ribonucleoside-diphosphate reductase large
PDBTitle: human ribonucleotide reductase r1 subunit (rrm1) in complex2 with datp and mg.
|
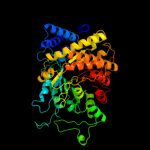
| 7 | c1xjeA_ |
|
 |
99.7 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ribonucleotide reductase, b12-dependent;
PDBTitle: structural mechanism of allosteric substrate specificity in a2 ribonucleotide reductase: dttp-gdp complex
|
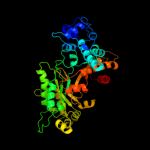
| 8 | c1pemA_ |
|
 |
99.6 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ribonucleoside-diphosphate reductase 2 alpha
PDBTitle: ribonucleotide reductase protein r1e from salmonella2 typhimurium
|
| 9 | d1r9da_ |
|
 |
99.5 |
18 |
Fold:PFL-like glycyl radical enzymes
Superfamily:PFL-like glycyl radical enzymes
Family:PFL-like |
| 10 | d1h16a_ |
|
 |
99.3 |
16 |
Fold:PFL-like glycyl radical enzymes
Superfamily:PFL-like glycyl radical enzymes
Family:PFL-like |
| 11 | c2f3oB_ |
|
 |
99.3 |
18 |
PDB header:unknown function
Chain: B: PDB Molecule:pyruvate formate-lyase 2;
PDBTitle: crystal structure of a glycyl radical enzyme from archaeoglobus2 fulgidus
|
| 12 | d1l1la_ |
|
 |
99.3 |
13 |
Fold:PFL-like glycyl radical enzymes
Superfamily:PFL-like glycyl radical enzymes
Family:B12-dependent (class II) ribonucleotide reductase |
| 13 | d1peqa2 |
|
 |
99.1 |
14 |
Fold:PFL-like glycyl radical enzymes
Superfamily:PFL-like glycyl radical enzymes
Family:R1 subunit of ribonucleotide reductase, C-terminal domain |
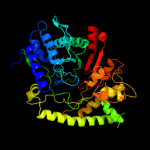
| 14 | c2y8nC_ |
|
 |
98.9 |
16 |
PDB header:lyase
Chain: C: PDB Molecule:4-hydroxyphenylacetate decarboxylase large subunit;
PDBTitle: crystal structure of glycyl radical enzyme
|
| 15 | d1rlra2 |
|
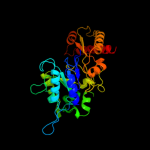 |
98.8 |
15 |
Fold:PFL-like glycyl radical enzymes
Superfamily:PFL-like glycyl radical enzymes
Family:R1 subunit of ribonucleotide reductase, C-terminal domain |
| 16 | d1qhma_ |
|
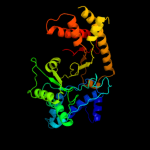 |
98.8 |
15 |
Fold:PFL-like glycyl radical enzymes
Superfamily:PFL-like glycyl radical enzymes
Family:PFL-like |
| 17 | d1rlra1 |
|
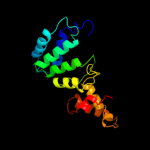 |
98.7 |
12 |
Fold:R1 subunit of ribonucleotide reductase, N-terminal domain
Superfamily:R1 subunit of ribonucleotide reductase, N-terminal domain
Family:R1 subunit of ribonucleotide reductase, N-terminal domain |
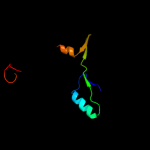
| 18 | c3a44D_ |
|
 |
96.0 |
17 |
PDB header:metal binding protein
Chain: D: PDB Molecule:hydrogenase nickel incorporation protein hypa;
PDBTitle: crystal structure of hypa in the dimeric form
|
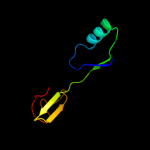
| 19 | c2kdxA_ |
|
 |
95.9 |
11 |
PDB header:metal-binding protein
Chain: A: PDB Molecule:hydrogenase/urease nickel incorporation protein
PDBTitle: solution structure of hypa protein
|
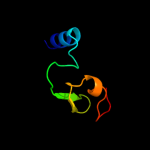
| 20 | c1dvbA_ |
|
 |
95.1 |
20 |
PDB header:electron transport
Chain: A: PDB Molecule:rubrerythrin;
PDBTitle: rubrerythrin
|
| 21 | c1yuzB_ |
|
not modelled |
94.1 |
26 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nigerythrin;
PDBTitle: partially reduced state of nigerythrin
|
| 22 | c2hr5B_ |
|
not modelled |
93.8 |
30 |
PDB header:metal binding protein
Chain: B: PDB Molecule:rubrerythrin;
PDBTitle: pf1283- rubrerythrin from pyrococcus furiosus iron bound form
|
| 23 | d2gmga1 |
|
not modelled |
89.2 |
18 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:PF0610-like |
| 24 | c2lcqA_ |
|
not modelled |
88.3 |
38 |
PDB header:metal binding protein
Chain: A: PDB Molecule:putative toxin vapc6;
PDBTitle: solution structure of the endonuclease nob1 from p.horikoshii
|
| 25 | d1lkoa2 |
|
not modelled |
85.6 |
33 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 26 | d1yuza2 |
|
not modelled |
85.1 |
29 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 27 | c3lpeF_ |
|
not modelled |
84.8 |
27 |
PDB header:transferase
Chain: F: PDB Molecule:dna-directed rna polymerase subunit e'';
PDBTitle: crystal structure of spt4/5ngn heterodimer complex from methanococcus2 jannaschii
|
| 28 | d1nnqa2 |
|
not modelled |
84.6 |
30 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 29 | d1ryqa_ |
|
not modelled |
82.5 |
28 |
Fold:Rubredoxin-like
Superfamily:RNA polymerase subunits
Family:RpoE2-like |
| 30 | d1dx8a_ |
|
not modelled |
82.2 |
40 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 31 | d2j0151 |
|
not modelled |
82.0 |
30 |
Fold:Rubredoxin-like
Superfamily:Zn-binding ribosomal proteins
Family:Ribosomal protein L32p |
| 32 | d2zjrz1 |
|
not modelled |
81.7 |
25 |
Fold:Rubredoxin-like
Superfamily:Zn-binding ribosomal proteins
Family:Ribosomal protein L32p |
| 33 | c1s24A_ |
|
not modelled |
81.6 |
38 |
PDB header:electron transport
Chain: A: PDB Molecule:rubredoxin 2;
PDBTitle: rubredoxin domain ii from pseudomonas oleovorans
|
| 34 | d1s24a_ |
|
not modelled |
81.6 |
38 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 35 | d2apob1 |
|
not modelled |
81.5 |
45 |
Fold:Rubredoxin-like
Superfamily:Nop10-like SnoRNP
Family:Nucleolar RNA-binding protein Nop10-like |
| 36 | d4rxna_ |
|
not modelled |
81.3 |
32 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 37 | c2v3bB_ |
|
not modelled |
81.2 |
35 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:rubredoxin 2;
PDBTitle: crystal structure of the electron transfer complex2 rubredoxin - rubredoxin reductase from pseudomonas3 aeruginosa.
|
| 38 | d1iroa_ |
|
not modelled |
81.0 |
32 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 39 | d1rb9a_ |
|
not modelled |
80.8 |
32 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 40 | d2ey4e1 |
|
not modelled |
80.4 |
40 |
Fold:Rubredoxin-like
Superfamily:Nop10-like SnoRNP
Family:Nucleolar RNA-binding protein Nop10-like |
| 41 | c2gb5B_ |
|
not modelled |
80.4 |
20 |
PDB header:hydrolase
Chain: B: PDB Molecule:nadh pyrophosphatase;
PDBTitle: crystal structure of nadh pyrophosphatase (ec 3.6.1.22) (1790429) from2 escherichia coli k12 at 2.30 a resolution
|
| 42 | d1h7va_ |
|
not modelled |
80.3 |
30 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 43 | d2gnra1 |
|
not modelled |
80.3 |
23 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:SSO2064-like |
| 44 | d1qcva_ |
|
not modelled |
80.3 |
19 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 45 | d1iu5a_ |
|
not modelled |
79.8 |
36 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 46 | c3axtA_ |
|
not modelled |
79.7 |
26 |
PDB header:transferase
Chain: A: PDB Molecule:probable n(2),n(2)-dimethylguanosine trna methyltransferase
PDBTitle: complex structure of trna methyltransferase trm1 from aquifex aeolicus2 with s-adenosyl-l-methionine
|
| 47 | d2dsxa1 |
|
not modelled |
79.2 |
36 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 48 | c2k2dA_ |
|
not modelled |
78.4 |
20 |
PDB header:metal binding protein
Chain: A: PDB Molecule:ring finger and chy zinc finger domain-
PDBTitle: solution nmr structure of c-terminal domain of human pirh2.2 northeast structural genomics consortium (nesg) target ht2c
|
| 49 | d1brfa_ |
|
not modelled |
76.9 |
36 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 50 | c2ba1B_ |
|
not modelled |
76.9 |
25 |
PDB header:rna binding protein
Chain: B: PDB Molecule:archaeal exosome rna binding protein csl4;
PDBTitle: archaeal exosome core
|
| 51 | d6rxna_ |
|
not modelled |
76.5 |
32 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 52 | d2cona1 |
|
not modelled |
73.9 |
35 |
Fold:Rubredoxin-like
Superfamily:NOB1 zinc finger-like
Family:NOB1 zinc finger-like |
| 53 | c2qkdA_ |
|
not modelled |
72.8 |
23 |
PDB header:signaling protein, cell cycle
Chain: A: PDB Molecule:zinc finger protein zpr1;
PDBTitle: crystal structure of tandem zpr1 domains
|
| 54 | c2nn6I_ |
|
not modelled |
72.6 |
13 |
PDB header:hydrolase/transferase
Chain: I: PDB Molecule:3'-5' exoribonuclease csl4 homolog;
PDBTitle: structure of the human rna exosome composed of rrp41, rrp45,2 rrp46, rrp43, mtr3, rrp42, csl4, rrp4, and rrp40
|
| 55 | d2rdva_ |
|
not modelled |
72.4 |
48 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 56 | d2fiya1 |
|
not modelled |
70.3 |
30 |
Fold:FdhE-like
Superfamily:FdhE-like
Family:FdhE-like |
| 57 | c2kn9A_ |
|
not modelled |
70.2 |
42 |
PDB header:electron transport
Chain: A: PDB Molecule:rubredoxin;
PDBTitle: solution structure of zinc-substituted rubredoxin b (rv3250c) from2 mycobacterium tuberculosis. seattle structural genomics center for3 infectious disease target mytud.01635.a
|
| 58 | c2f9iD_ |
|
not modelled |
69.1 |
22 |
PDB header:transferase
Chain: D: PDB Molecule:acetyl-coenzyme a carboxylase carboxyl
PDBTitle: crystal structure of the carboxyltransferase subunit of acc2 from staphylococcus aureus
|
| 59 | d2ct7a1 |
|
not modelled |
66.0 |
28 |
Fold:RING/U-box
Superfamily:RING/U-box
Family:IBR domain |
| 60 | d2qam01 |
|
not modelled |
65.2 |
23 |
Fold:Rubredoxin-like
Superfamily:Zn-binding ribosomal proteins
Family:Ribosomal protein L32p |
| 61 | c1u5kA_ |
|
not modelled |
65.0 |
19 |
PDB header:recombination,replication
Chain: A: PDB Molecule:hypothetical protein;
PDBTitle: recombinational repair protein reco
|
| 62 | d2dkta1 |
|
not modelled |
63.6 |
24 |
Fold:CHY zinc finger-like
Superfamily:CHY zinc finger-like
Family:CHY zinc finger |
| 63 | d1pfva3 |
|
not modelled |
62.7 |
30 |
Fold:Rubredoxin-like
Superfamily:Methionyl-tRNA synthetase (MetRS), Zn-domain
Family:Methionyl-tRNA synthetase (MetRS), Zn-domain |
| 64 | c3ir9A_ |
|
not modelled |
62.2 |
35 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:peptide chain release factor subunit 1;
PDBTitle: c-terminal domain of peptide chain release factor from2 methanosarcina mazei.
|
| 65 | c2ktvA_ |
|
not modelled |
61.3 |
19 |
PDB header:translation
Chain: A: PDB Molecule:eukaryotic peptide chain release factor subunit 1;
PDBTitle: human erf1 c-domain, "open" conformer
|
| 66 | d1nuia2 |
|
not modelled |
60.5 |
36 |
Fold:Rubredoxin-like
Superfamily:Zinc beta-ribbon
Family:DNA primase zinc finger |
| 67 | c2f9yB_ |
|
not modelled |
59.8 |
32 |
PDB header:ligase
Chain: B: PDB Molecule:acetyl-coenzyme a carboxylase carboxyl transferase subunit
PDBTitle: the crystal structure of the carboxyltransferase subunit of acc from2 escherichia coli
|
| 68 | d2f9yb1 |
|
not modelled |
59.8 |
32 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Biotin dependent carboxylase carboxyltransferase domain |
| 69 | d1ltla_ |
|
not modelled |
59.6 |
22 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:DNA replication initiator (cdc21/cdc54) N-terminal domain |
| 70 | c2aklA_ |
|
not modelled |
56.1 |
21 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:phna-like protein pa0128;
PDBTitle: solution structure for phn-a like protein pa0128 from2 pseudomonas aeruginosa
|
| 71 | c3f2cA_ |
|
not modelled |
56.0 |
40 |
PDB header:transferase/dna
Chain: A: PDB Molecule:geobacillus kaustophilus dna polc;
PDBTitle: dna polymerase polc from geobacillus kaustophilus complex with dna,2 dgtp and mn
|
| 72 | d2nn6i1 |
|
not modelled |
55.0 |
13 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 73 | c3cngC_ |
|
not modelled |
54.5 |
29 |
PDB header:hydrolase
Chain: C: PDB Molecule:nudix hydrolase;
PDBTitle: crystal structure of nudix hydrolase from nitrosomonas europaea
|
| 74 | c1ltlE_ |
|
not modelled |
54.1 |
22 |
PDB header:replication
Chain: E: PDB Molecule:dna replication initiator (cdc21/cdc54);
PDBTitle: the dodecamer structure of mcm from archaeal m.2 thermoautotrophicum
|
| 75 | c3bjiA_ |
|
not modelled |
53.7 |
38 |
PDB header:signaling protein
Chain: A: PDB Molecule:proto-oncogene vav;
PDBTitle: structural basis of promiscuous guanine nucleotide exchange2 by the t-cell essential vav1
|
| 76 | d2akla2 |
|
not modelled |
53.6 |
23 |
Fold:Rubredoxin-like
Superfamily:Zinc beta-ribbon
Family:PhnA zinc-binding domain |
| 77 | c1m2oA_ |
|
not modelled |
53.0 |
13 |
PDB header:protein transport/signaling protein
Chain: A: PDB Molecule:protein transport protein sec23;
PDBTitle: crystal structure of the sec23-sar1 complex
|
| 78 | c2vl6C_ |
|
not modelled |
51.4 |
30 |
PDB header:dna binding protein
Chain: C: PDB Molecule:minichromosome maintenance protein mcm;
PDBTitle: structural analysis of the sulfolobus solfataricus mcm2 protein n-terminal domain
|
| 79 | c1vddC_ |
|
not modelled |
51.4 |
19 |
PDB header:recombination
Chain: C: PDB Molecule:recombination protein recr;
PDBTitle: crystal structure of recombinational repair protein recr
|
| 80 | d1pfta_ |
|
not modelled |
51.2 |
25 |
Fold:Rubredoxin-like
Superfamily:Zinc beta-ribbon
Family:Transcriptional factor domain |
| 81 | c2k5cA_ |
|
not modelled |
48.1 |
56 |
PDB header:metal binding protein
Chain: A: PDB Molecule:uncharacterized protein pf0385;
PDBTitle: nmr structure for pf0385
|
| 82 | d1u5ka2 |
|
not modelled |
48.0 |
19 |
Fold:ArfGap/RecO-like zinc finger
Superfamily:ArfGap/RecO-like zinc finger
Family:RecO C-terminal domain-like |
| 83 | d1dl6a_ |
|
not modelled |
47.9 |
29 |
Fold:Rubredoxin-like
Superfamily:Zinc beta-ribbon
Family:Transcriptional factor domain |
| 84 | d2dlqa2 |
|
not modelled |
47.0 |
75 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 85 | c3ky9B_ |
|
not modelled |
47.0 |
38 |
PDB header:apoptosis
Chain: B: PDB Molecule:proto-oncogene vav;
PDBTitle: autoinhibited vav1
|
| 86 | d1ynjd1 |
|
not modelled |
44.9 |
35 |
Fold:beta and beta-prime subunits of DNA dependent RNA-polymerase
Superfamily:beta and beta-prime subunits of DNA dependent RNA-polymerase
Family:RNA-polymerase beta-prime |
| 87 | c3gn5B_ |
|
not modelled |
44.5 |
36 |
PDB header:dna binding protein
Chain: B: PDB Molecule:hth-type transcriptional regulator mqsa (ygit/b3021);
PDBTitle: structure of the e. coli protein mqsa (ygit/b3021)
|
| 88 | c3eswA_ |
|
not modelled |
43.4 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptide-n(4)-(n-acetyl-beta-glucosaminyl)asparagine
PDBTitle: complex of yeast pngase with glcnac2-iac.
|
| 89 | d2ctda2 |
|
not modelled |
43.3 |
50 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 90 | d1x6ma_ |
|
not modelled |
42.6 |
28 |
Fold:Mss4-like
Superfamily:Mss4-like
Family:Glutathione-dependent formaldehyde-activating enzyme, Gfa |
| 91 | c1ee8A_ |
|
not modelled |
42.5 |
26 |
PDB header:dna binding protein
Chain: A: PDB Molecule:mutm (fpg) protein;
PDBTitle: crystal structure of mutm (fpg) protein from thermus thermophilus hb8
|
| 92 | c3eh2B_ |
|
not modelled |
42.5 |
19 |
PDB header:protein transport
Chain: B: PDB Molecule:protein transport protein sec24c;
PDBTitle: crystal structure of the human copii-coat protein sec24c
|
| 93 | c1m2vB_ |
|
not modelled |
42.2 |
20 |
PDB header:protein transport
Chain: B: PDB Molecule:protein transport protein sec24;
PDBTitle: crystal structure of the yeast sec23/24 heterodimer
|
| 94 | c2owoA_ |
|
not modelled |
41.0 |
44 |
PDB header:ligase/dna
Chain: A: PDB Molecule:dna ligase;
PDBTitle: last stop on the road to repair: structure of e.coli dna ligase bound2 to nicked dna-adenylate
|
| 95 | c3k7aM_ |
|
not modelled |
40.3 |
29 |
PDB header:transcription
Chain: M: PDB Molecule:transcription initiation factor iib;
PDBTitle: crystal structure of an rna polymerase ii-tfiib complex
|
| 96 | c3h0gN_ |
|
not modelled |
40.0 |
21 |
PDB header:transcription
Chain: N: PDB Molecule:dna-directed rna polymerase ii subunit rpb2;
PDBTitle: rna polymerase ii from schizosaccharomyces pombe
|
| 97 | c2ysaA_ |
|
not modelled |
39.7 |
21 |
PDB header:metal binding protein
Chain: A: PDB Molecule:retinoblastoma-binding protein 6;
PDBTitle: solution structure of the zinc finger cchc domain from the2 human retinoblastoma-binding protein 6 (retinoblastoma-3 binding q protein 1, rbq-1)
|
| 98 | c3cw2M_ |
|
not modelled |
39.2 |
22 |
PDB header:translation
Chain: M: PDB Molecule:translation initiation factor 2 subunit beta;
PDBTitle: crystal structure of the intact archaeal translation2 initiation factor 2 from sulfolobus solfataricus .
|
| 99 | c1pd0A_ |
|
not modelled |
39.2 |
20 |
PDB header:transport protein
Chain: A: PDB Molecule:protein transport protein sec24;
PDBTitle: crystal structure of the copii coat subunit, sec24,2 complexed with a peptide from the snare protein sed53 (yeast syntaxin-5)
|
| 100 | d1vdda_ |
|
not modelled |
39.1 |
23 |
Fold:Recombination protein RecR
Superfamily:Recombination protein RecR
Family:Recombination protein RecR |
| 101 | c2opfA_ |
|
not modelled |
38.5 |
33 |
PDB header:hydrolase/dna
Chain: A: PDB Molecule:endonuclease viii;
PDBTitle: crystal structure of the dna repair enzyme endonuclease-viii (nei)2 from e. coli (r252a) in complex with ap-site containing dna substrate
|
| 102 | c3eg9B_ |
|
not modelled |
38.4 |
16 |
PDB header:protein transport
Chain: B: PDB Molecule:sec24 related gene family, member d;
PDBTitle: crystal structure of the mammalian copii-coat protein2 sec23/24 bound to the transport signal sequence of membrin
|
| 103 | c2yqqA_ |
|
not modelled |
38.1 |
29 |
PDB header:protein binding
Chain: A: PDB Molecule:zinc finger hit domain-containing protein 3;
PDBTitle: solution structure of the zf-hit domain in zinc finger hit2 domain-containing protein 3 (trip-3)
|
| 104 | d1j6wa_ |
|
not modelled |
38.1 |
57 |
Fold:LuxS/MPP-like metallohydrolase
Superfamily:LuxS/MPP-like metallohydrolase
Family:Autoinducer-2 production protein LuxS |
| 105 | c3h7hA_ |
|
not modelled |
38.1 |
19 |
PDB header:transcription
Chain: A: PDB Molecule:transcription elongation factor spt4;
PDBTitle: crystal structure of the human transcription elongation factor dsif,2 hspt4/hspt5 (176-273)
|
| 106 | c2pmzB_ |
|
not modelled |
38.0 |
28 |
PDB header:translation, transferase
Chain: B: PDB Molecule:dna-directed rna polymerase subunit b;
PDBTitle: archaeal rna polymerase from sulfolobus solfataricus
|
| 107 | c2ja6L_ |
|
not modelled |
37.7 |
21 |
PDB header:transferase
Chain: L: PDB Molecule:dna-directed rna polymerases i, ii, and iii 7.7
PDBTitle: cpd lesion containing rna polymerase ii elongation complex2 b
|
| 108 | c2bx9J_ |
|
not modelled |
37.3 |
35 |
PDB header:transcription regulation
Chain: J: PDB Molecule:tryptophan rna-binding attenuator protein-inhibitory
PDBTitle: crystal structure of b.subtilis anti-trap protein, an2 antagonist of trap-rna interactions
|
| 109 | c3na7A_ |
|
not modelled |
37.3 |
20 |
PDB header:gene regulation, chaperone
Chain: A: PDB Molecule:hp0958;
PDBTitle: 2.2 angstrom structure of the hp0958 protein from helicobacter pylori2 ccug 17874
|
| 110 | c3h0gL_ |
|
not modelled |
36.8 |
21 |
PDB header:transcription
Chain: L: PDB Molecule:dna-directed rna polymerases i, ii, and iii
PDBTitle: rna polymerase ii from schizosaccharomyces pombe
|
| 111 | d1x3za1 |
|
not modelled |
36.7 |
22 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:Transglutaminase core |
| 112 | c1nnjA_ |
|
not modelled |
36.6 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:formamidopyrimidine-dna glycosylase;
PDBTitle: crystal structure complex between the lactococcus lactis fpg and an2 abasic site containing dna
|
| 113 | c2f5qA_ |
|
not modelled |
35.9 |
32 |
PDB header:hydrolase/dna
Chain: A: PDB Molecule:formamidopyrimidine-dna glycosidase;
PDBTitle: catalytically inactive (e3q) mutm crosslinked to oxog:c2 containing dna cc2
|
| 114 | c3pkiF_ |
|
not modelled |
35.5 |
28 |
PDB header:hydrolase
Chain: F: PDB Molecule:nad-dependent deacetylase sirtuin-6;
PDBTitle: human sirt6 crystal structure in complex with adp ribose
|
| 115 | d1j6xa_ |
|
not modelled |
35.3 |
43 |
Fold:LuxS/MPP-like metallohydrolase
Superfamily:LuxS/MPP-like metallohydrolase
Family:Autoinducer-2 production protein LuxS |
| 116 | c1v9pB_ |
|
not modelled |
35.3 |
56 |
PDB header:ligase
Chain: B: PDB Molecule:dna ligase;
PDBTitle: crystal structure of nad+-dependent dna ligase
|
| 117 | d2dula1 |
|
not modelled |
35.1 |
17 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:TRM1-like |
| 118 | d1srka_ |
|
not modelled |
35.0 |
33 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 119 | c3pfqA_ |
|
not modelled |
35.0 |
29 |
PDB header:transferase
Chain: A: PDB Molecule:protein kinase c beta type;
PDBTitle: crystal structure and allosteric activation of protein kinase c beta2 ii
|
| 120 | c3f9vA_ |
|
not modelled |
34.9 |
30 |
PDB header:hydrolase
Chain: A: PDB Molecule:minichromosome maintenance protein mcm;
PDBTitle: crystal structure of a near full-length archaeal mcm: functional2 insights for an aaa+ hexameric helicase
|

