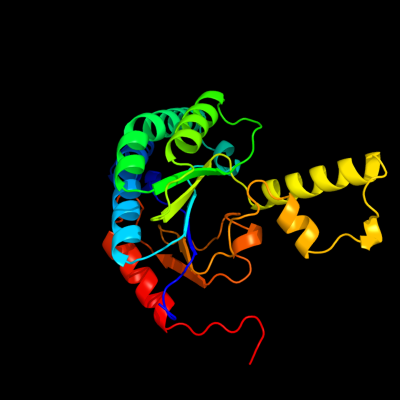
| 1 | c2vg2C_
|
|
 |
100.0 |
41 |
PDB header:transferase
Chain: C: PDB Molecule:undecaprenyl pyrophosphate synthetase;
PDBTitle: rv2361 with ipp
|
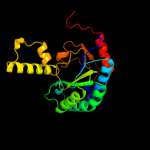
| 2 | d1ueha_
|
|
 |
100.0 |
97 |
Fold:Undecaprenyl diphosphate synthase
Superfamily:Undecaprenyl diphosphate synthase
Family:Undecaprenyl diphosphate synthase |
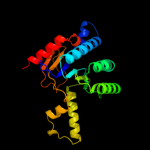
| 3 | c2vfwB_
|
|
 |
100.0 |
33 |
PDB header:transferase
Chain: B: PDB Molecule:short-chain z-isoprenyl diphosphate synthetase;
PDBTitle: rv1086 native
|
| 4 | c1jp3A_
|
|
 |
100.0 |
95 |
PDB header:transferase
Chain: A: PDB Molecule:undecaprenyl pyrophosphate synthase;
PDBTitle: structure of e.coli undecaprenyl pyrophosphate synthase
|
| 5 | d1f75a_
|
|
 |
100.0 |
38 |
Fold:Undecaprenyl diphosphate synthase
Superfamily:Undecaprenyl diphosphate synthase
Family:Undecaprenyl diphosphate synthase |
| 6 | c2d2rA_
|
|
 |
100.0 |
41 |
PDB header:transferase
Chain: A: PDB Molecule:undecaprenyl pyrophosphate synthase;
PDBTitle: crystal structure of helicobacter pylori undecaprenyl pyrophosphate2 synthase
|
| 7 | c3ugsB_
|
|
 |
100.0 |
38 |
PDB header:transferase
Chain: B: PDB Molecule:undecaprenyl pyrophosphate synthase;
PDBTitle: crystal structure of a probable undecaprenyl diphosphate synthase2 (upps) from campylobacter jejuni
|
| 8 | c3bdkB_
|
|
 |
92.3 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:d-mannonate dehydratase;
PDBTitle: crystal structure of streptococcus suis mannonate2 dehydratase complexed with substrate analogue
|
| 9 | c3cnyA_
|
|
 |
91.4 |
8 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:inositol catabolism protein iole;
PDBTitle: crystal structure of a putative inositol catabolism protein iole2 (iole, lp_3607) from lactobacillus plantarum wcfs1 at 1.85 a3 resolution
|
| 10 | d1tz9a_
|
|
 |
85.7 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:UxuA-like |
| 11 | c2ou4C_
|
|
 |
84.2 |
9 |
PDB header:isomerase
Chain: C: PDB Molecule:d-tagatose 3-epimerase;
PDBTitle: crystal structure of d-tagatose 3-epimerase from2 pseudomonas cichorii
|
| 12 | c2hk1D_
|
|
 |
84.1 |
12 |
PDB header:isomerase
Chain: D: PDB Molecule:d-psicose 3-epimerase;
PDBTitle: crystal structure of d-psicose 3-epimerase (dpease) in the presence of2 d-fructose
|
| 13 | c2nuxB_
|
|
 |
81.3 |
9 |
PDB header:lyase
Chain: B: PDB Molecule:2-keto-3-deoxygluconate/2-keto-3-deoxy-6-phospho gluconate
PDBTitle: 2-keto-3-deoxygluconate aldolase from sulfolobus acidocaldarius,2 native structure in p6522 at 2.5 a resolution
|
| 14 | c3dx5A_
|
|
 |
79.6 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:uncharacterized protein asbf;
PDBTitle: crystal structure of the probable 3-dhs dehydratase asbf involved in2 the petrobactin synthesis from bacillus anthracis
|
| 15 | c3eb2A_
|
|
 |
77.4 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:putative dihydrodipicolinate synthetase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 2.0a resolution
|
| 16 | c3b4uB_
|
|
 |
75.9 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from agrobacterium2 tumefaciens str. c58
|
| 17 | c3fluD_
|
|
 |
75.3 |
20 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from the pathogen2 neisseria meningitidis
|
| 18 | c2rfgB_
|
|
 |
73.9 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from hahella2 chejuensis at 1.5a resolution
|
| 19 | c3cqkB_
|
|
 |
73.5 |
16 |
PDB header:isomerase
Chain: B: PDB Molecule:l-ribulose-5-phosphate 3-epimerase ulae;
PDBTitle: crystal structure of l-xylulose-5-phosphate 3-epimerase ulae (form b)2 complex with zn2+ and sulfate
|
| 20 | c3d0cB_
|
|
 |
73.4 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 oceanobacillus iheyensis at 1.9 a resolution
|
| 21 | c3qxbB_ |
|
not modelled |
72.1 |
9 |
PDB header:isomerase
Chain: B: PDB Molecule:putative xylose isomerase;
PDBTitle: crystal structure of a putative xylose isomerase (yp_426450.1) from2 rhodospirillum rubrum atcc 11170 at 1.90 a resolution
|
| 22 | c3si9B_ |
|
not modelled |
70.7 |
19 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from bartonella2 henselae
|
| 23 | c2zdsB_ |
|
not modelled |
66.6 |
15 |
PDB header:dna binding protein
Chain: B: PDB Molecule:putative dna-binding protein;
PDBTitle: crystal structure of sco6571 from streptomyces coelicolor2 a3(2)
|
| 24 | c3pueA_ |
|
not modelled |
63.5 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of the complex of dhydrodipicolinate synthase from2 acinetobacter baumannii with lysine at 2.6a resolution
|
| 25 | c3s5oA_ |
|
not modelled |
59.4 |
10 |
PDB header:lyase
Chain: A: PDB Molecule:4-hydroxy-2-oxoglutarate aldolase, mitochondrial;
PDBTitle: crystal structure of human 4-hydroxy-2-oxoglutarate aldolase bound to2 pyruvate
|
| 26 | c2r8wB_ |
|
not modelled |
57.4 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:agr_c_1641p;
PDBTitle: the crystal structure of dihydrodipicolinate synthase (atu0899) from2 agrobacterium tumefaciens str. c58
|
| 27 | c3cprB_ |
|
not modelled |
56.4 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthetase;
PDBTitle: the crystal structure of corynebacterium glutamicum2 dihydrodipicolinate synthase to 2.2 a resolution
|
| 28 | d1bxba_ |
|
not modelled |
55.5 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 29 | c2v9dB_ |
|
not modelled |
54.9 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:yage;
PDBTitle: crystal structure of yage, a prophage protein belonging to2 the dihydrodipicolinic acid synthase family from e. coli3 k12
|
| 30 | c3fkkA_ |
|
not modelled |
52.6 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:l-2-keto-3-deoxyarabonate dehydratase;
PDBTitle: structure of l-2-keto-3-deoxyarabonate dehydratase
|
| 31 | c3kwsB_ |
|
not modelled |
52.3 |
3 |
PDB header:isomerase
Chain: B: PDB Molecule:putative sugar isomerase;
PDBTitle: crystal structure of putative sugar isomerase (yp_001305149.1) from2 parabacteroides distasonis atcc 8503 at 1.68 a resolution
|
| 32 | d1m5wa_ |
|
not modelled |
51.6 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Pyridoxine 5'-phosphate synthase
Family:Pyridoxine 5'-phosphate synthase |
| 33 | c3qfeB_ |
|
not modelled |
51.3 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:putative dihydrodipicolinate synthase family protein;
PDBTitle: crystal structures of a putative dihydrodipicolinate synthase family2 protein from coccidioides immitis
|
| 34 | c3noeA_ |
|
not modelled |
49.5 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from pseudomonas2 aeruginosa
|
| 35 | d1xxxa1 |
|
not modelled |
49.2 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 36 | c3n2xB_ |
|
not modelled |
48.4 |
18 |
PDB header:lyase
Chain: B: PDB Molecule:uncharacterized protein yage;
PDBTitle: crystal structure of yage, a prophage protein belonging to the2 dihydrodipicolinic acid synthase family from e. coli k12 in complex3 with pyruvate
|
| 37 | c3ktcB_ |
|
not modelled |
46.8 |
8 |
PDB header:isomerase
Chain: B: PDB Molecule:xylose isomerase;
PDBTitle: crystal structure of putative sugar isomerase (yp_050048.1) from2 erwinia carotovora atroseptica scri1043 at 1.54 a resolution
|
| 38 | c2ehhE_ |
|
not modelled |
46.6 |
16 |
PDB header:lyase
Chain: E: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 aquifex aeolicus
|
| 39 | c3h5dD_ |
|
not modelled |
46.6 |
13 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: dihydrodipicolinate synthase from drug-resistant streptococcus2 pneumoniae
|
| 40 | c2vc6A_ |
|
not modelled |
45.3 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: structure of mosa from s. meliloti with pyruvate bound
|
| 41 | c3bi8A_ |
|
not modelled |
44.1 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: structure of dihydrodipicolinate synthase from clostridium2 botulinum
|
| 42 | c2r94B_ |
|
not modelled |
43.7 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:2-keto-3-deoxy-(6-phospho-)gluconate aldolase;
PDBTitle: crystal structure of kd(p)ga from t.tenax
|
| 43 | c2zvrA_ |
|
not modelled |
42.9 |
9 |
PDB header:isomerase
Chain: A: PDB Molecule:uncharacterized protein tm_0416;
PDBTitle: crystal structure of a d-tagatose 3-epimerase-related2 protein from thermotoga maritima
|
| 44 | c2ksnA_ |
|
not modelled |
41.9 |
20 |
PDB header:signaling protein
Chain: A: PDB Molecule:ubiquitin domain-containing protein 2;
PDBTitle: solution structure of the n-terminal domain of dc-ubp/ubtd2
|
| 45 | d1hl2a_ |
|
not modelled |
41.9 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 46 | d1vhua_ |
|
not modelled |
41.7 |
15 |
Fold:Macro domain-like
Superfamily:Macro domain-like
Family:Macro domain |
| 47 | d1i60a_ |
|
not modelled |
41.6 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:IolI-like |
| 48 | d1bxca_ |
|
not modelled |
39.3 |
7 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 49 | c3daqB_ |
|
not modelled |
38.9 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from methicillin-2 resistant staphylococcus aureus
|
| 50 | d1otha2 |
|
not modelled |
37.7 |
21 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 51 | c2ejaB_ |
|
not modelled |
37.6 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: crystal structure of uroporphyrinogen decarboxylase from2 aquifex aeolicus
|
| 52 | d1o5ka_ |
|
not modelled |
37.5 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 53 | c3g0sA_ |
|
not modelled |
36.0 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: dihydrodipicolinate synthase from salmonella typhimurium lt2
|
| 54 | c3na8A_ |
|
not modelled |
35.9 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:putative dihydrodipicolinate synthetase;
PDBTitle: crystal structure of a putative dihydrodipicolinate synthetase from2 pseudomonas aeruginosa
|
| 55 | c3dz1A_ |
|
not modelled |
35.7 |
22 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 1.87a resolution
|
| 56 | c3lciA_ |
|
not modelled |
34.7 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:n-acetylneuraminate lyase;
PDBTitle: the d-sialic acid aldolase mutant v251w
|
| 57 | c3bzjA_ |
|
not modelled |
33.3 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:uv endonuclease;
PDBTitle: uvde k229l
|
| 58 | c1xhoB_ |
|
not modelled |
33.2 |
19 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:chorismate mutase;
PDBTitle: chorismate mutase from clostridium thermocellum cth-682
|
| 59 | d1xhoa_ |
|
not modelled |
33.2 |
19 |
Fold:Bacillus chorismate mutase-like
Superfamily:YjgF-like
Family:Chorismate mutase |
| 60 | c3vh1A_ |
|
not modelled |
31.8 |
20 |
PDB header:metal binding protein
Chain: A: PDB Molecule:ubiquitin-like modifier-activating enzyme atg7;
PDBTitle: crystal structure of saccharomyces cerevisiae atg7 (1-595)
|
| 61 | c3gk0H_ |
|
not modelled |
31.4 |
15 |
PDB header:transferase
Chain: H: PDB Molecule:pyridoxine 5'-phosphate synthase;
PDBTitle: crystal structure of pyridoxal phosphate biosynthetic2 protein from burkholderia pseudomallei
|
| 62 | d1fnja_ |
|
not modelled |
31.2 |
22 |
Fold:Bacillus chorismate mutase-like
Superfamily:YjgF-like
Family:Chorismate mutase |
| 63 | c3q71A_ |
|
not modelled |
30.9 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:poly [adp-ribose] polymerase 14;
PDBTitle: human parp14 (artd8) - macro domain 2 in complex with adenosine-5-2 diphosphoribose
|
| 64 | c3grfA_ |
|
not modelled |
30.8 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:ornithine carbamoyltransferase;
PDBTitle: x-ray structure of ornithine transcarbamoylase from giardia2 lamblia
|
| 65 | d1vk1a_ |
|
not modelled |
29.8 |
60 |
Fold:ParB/Sulfiredoxin
Superfamily:ParB/Sulfiredoxin
Family:Hypothetical protein PF0380 |
| 66 | c1ut8B_ |
|
not modelled |
28.1 |
8 |
PDB header:hydrolase
Chain: B: PDB Molecule:exodeoxyribonuclease;
PDBTitle: divalent metal ions (zinc) bound to t5 5'-exonuclease
|
| 67 | c2yxgD_ |
|
not modelled |
27.7 |
18 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihyrodipicolinate synthase (dapa)
|
| 68 | d1dbfa_ |
|
not modelled |
25.6 |
22 |
Fold:Bacillus chorismate mutase-like
Superfamily:YjgF-like
Family:Chorismate mutase |
| 69 | c2pfuA_ |
|
not modelled |
25.6 |
10 |
PDB header:transport protein
Chain: A: PDB Molecule:biopolymer transport exbd protein;
PDBTitle: nmr strcuture determination of the periplasmic domain of exbd from2 e.coli
|
| 70 | c2f9iC_ |
|
not modelled |
25.5 |
16 |
PDB header:transferase
Chain: C: PDB Molecule:acetyl-coenzyme a carboxylase carboxyl
PDBTitle: crystal structure of the carboxyltransferase subunit of acc2 from staphylococcus aureus
|
| 71 | c3gzaB_ |
|
not modelled |
24.6 |
16 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative alpha-l-fucosidase;
PDBTitle: crystal structure of putative alpha-l-fucosidase (np_812709.1) from2 bacteroides thetaiotaomicron vpi-5482 at 1.60 a resolution
|
| 72 | d1cmwa2 |
|
not modelled |
24.5 |
10 |
Fold:PIN domain-like
Superfamily:PIN domain-like
Family:5' to 3' exonuclease catalytic domain |
| 73 | d1xkya1 |
|
not modelled |
23.2 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 74 | d2f9ya1 |
|
not modelled |
22.9 |
16 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Biotin dependent carboxylase carboxyltransferase domain |
| 75 | d2a6na1 |
|
not modelled |
22.7 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 76 | c3lmzA_ |
|
not modelled |
22.5 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:putative sugar isomerase;
PDBTitle: crystal structure of putative sugar isomerase. (yp_001305105.1) from2 parabacteroides distasonis atcc 8503 at 1.44 a resolution
|
| 77 | c3e96B_ |
|
not modelled |
22.4 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 bacillus clausii
|
| 78 | c3ngfA_ |
|
not modelled |
22.3 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:ap endonuclease, family 2;
PDBTitle: crystal structure of ap endonuclease, family 2 from brucella2 melitensis
|
| 79 | d1nw9b_ |
|
not modelled |
21.7 |
16 |
Fold:Caspase-like
Superfamily:Caspase-like
Family:Caspase catalytic domain |
| 80 | c3ju2A_ |
|
not modelled |
21.6 |
24 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein smc04130;
PDBTitle: crystal structure of protein smc04130 from sinorhizobium meliloti 1021
|
| 81 | d1xima_ |
|
not modelled |
21.5 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 82 | c2qtzA_ |
|
not modelled |
20.6 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:methionine synthase reductase;
PDBTitle: crystal structure of the nadp+-bound fad-containing fnr-like module of2 human methionine synthase reductase
|
| 83 | c3gqeA_ |
|
not modelled |
19.4 |
13 |
PDB header:viral protein
Chain: A: PDB Molecule:non-structural protein 3;
PDBTitle: crystal structure of macro domain of venezuelan equine encephalitis2 virus
|
| 84 | c3q6zA_ |
|
not modelled |
16.3 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:poly [adp-ribose] polymerase 14;
PDBTitle: human parp14 (artd8)-macro domain 1 in complex with adenosine-5-2 diphosphoribose
|
| 85 | d1f1ja_ |
|
not modelled |
16.1 |
16 |
Fold:Caspase-like
Superfamily:Caspase-like
Family:Caspase catalytic domain |
| 86 | d1fmta2 |
|
not modelled |
16.1 |
17 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
| 87 | d2eyqa6 |
|
not modelled |
15.9 |
9 |
Fold:TRCF domain-like
Superfamily:TRCF domain-like
Family:TRCF domain |
| 88 | d1wsca1 |
|
not modelled |
15.7 |
16 |
Fold:AMMECR1-like
Superfamily:AMMECR1-like
Family:AMMECR1-like |
| 89 | c2bpoA_ |
|
not modelled |
15.6 |
9 |
PDB header:reductase
Chain: A: PDB Molecule:nadph-cytochrom p450 reductase;
PDBTitle: crystal structure of the yeast cpr triple mutant: d74g,2 y75f, k78a.
|
| 90 | c1nmqB_ |
|
not modelled |
15.6 |
20 |
PDB header:apoptosis, hydrolase
Chain: B: PDB Molecule:caspase-3;
PDBTitle: extendend tethering: in situ assembly of inhibitors
|
| 91 | c3lerA_ |
|
not modelled |
15.5 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 campylobacter jejuni subsp. jejuni nctc 11168
|
| 92 | d1w3ia_ |
|
not modelled |
15.2 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 93 | c2btdA_ |
|
not modelled |
15.0 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:pts-dependent dihydroxyacetone kinase;
PDBTitle: crystal structure of dhal from e. coli
|
| 94 | d1bg4a_ |
|
not modelled |
14.9 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 95 | c3eyxB_ |
|
not modelled |
14.5 |
36 |
PDB header:lyase
Chain: B: PDB Molecule:carbonic anhydrase;
PDBTitle: crystal structure of carbonic anhydrase nce103 from2 saccharomyces cerevisiae
|
| 96 | d2pp4a1 |
|
not modelled |
14.4 |
13 |
Fold:TAFH domain-like
Superfamily:TAFH domain-like
Family:TAFH domain-like |
| 97 | d1yvka1 |
|
not modelled |
14.4 |
17 |
Fold:Acyl-CoA N-acyltransferases (Nat)
Superfamily:Acyl-CoA N-acyltransferases (Nat)
Family:N-acetyl transferase, NAT |
| 98 | c2x7vA_ |
|
not modelled |
14.3 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:probable endonuclease 4;
PDBTitle: crystal structure of thermotoga maritima endonuclease iv in2 the presence of zinc
|
| 99 | d2q02a1 |
|
not modelled |
13.7 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:IolI-like |