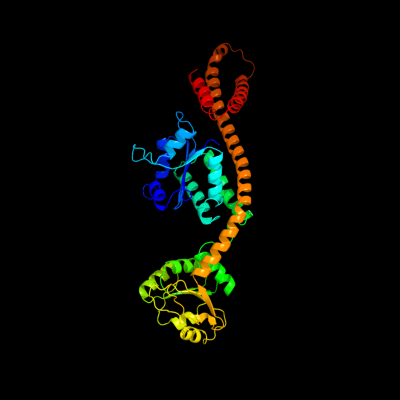
| 1 | c1gpjA_
|
|
 |
100.0 |
34 |
PDB header:reductase
Chain: A: PDB Molecule:glutamyl-trna reductase;
PDBTitle: glutamyl-trna reductase from methanopyrus kandleri
|
| 2 | d1gpja3
|
|
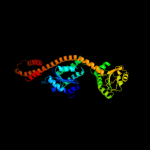
 |
100.0 |
37 |
Fold:Ferredoxin-like
Superfamily:Glutamyl tRNA-reductase catalytic, N-terminal domain
Family:Glutamyl tRNA-reductase catalytic, N-terminal domain |
| 3 | d1gpja2
|
|
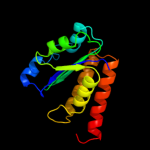
 |
100.0 |
40 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
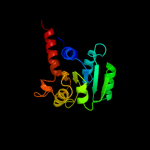
| 4 | c3oj0A_
|
|
 |
99.9 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glutamyl-trna reductase;
PDBTitle: crystal structure of glutamyl-trna reductase from thermoplasma2 volcanium (nucleotide binding domain)
|
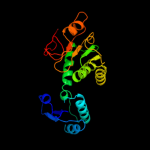
| 5 | c1luaA_
|
|
 |
99.8 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:methylene tetrahydromethanopterin dehydrogenase;
PDBTitle: structure of methylene-tetrahydromethanopterin dehydrogenase from2 methylobacterium extorquens am1 complexed with nadp
|
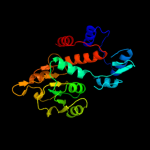
| 6 | c3pgjB_
|
|
 |
99.8 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:shikimate dehydrogenase;
PDBTitle: 2.49 angstrom resolution crystal structure of shikimate 5-2 dehydrogenase (aroe) from vibrio cholerae o1 biovar eltor str. n169613 in complex with shikimate
|
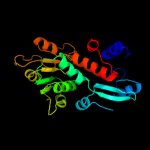
| 7 | c2nloA_
|
|
 |
99.7 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:shikimate dehydrogenase;
PDBTitle: crystal structure of the quinate dehydrogenase from corynebacterium2 glutamicum
|
| 8 | d1gpja1
|
|
 |
99.7 |
15 |
Fold:Glutamyl tRNA-reductase dimerization domain
Superfamily:Glutamyl tRNA-reductase dimerization domain
Family:Glutamyl tRNA-reductase dimerization domain |
| 9 | c3donA_
|
|
 |
99.7 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:shikimate dehydrogenase;
PDBTitle: crystal structure of shikimate dehydrogenase from staphylococcus2 epidermidis
|
| 10 | c2eggA_
|
|
 |
99.7 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:shikimate 5-dehydrogenase;
PDBTitle: crystal structure of shikimate 5-dehydrogenase (aroe) from2 geobacillus kaustophilus
|
| 11 | c3gvpB_
|
|
 |
99.6 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:adenosylhomocysteinase 3;
PDBTitle: human sahh-like domain of human adenosylhomocysteinase 3
|
| 12 | c1lehB_
|
|
 |
99.6 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:leucine dehydrogenase;
PDBTitle: leucine dehydrogenase from bacillus sphaericus
|
| 13 | c3o8qB_
|
|
 |
99.6 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:shikimate 5-dehydrogenase i alpha;
PDBTitle: 1.45 angstrom resolution crystal structure of shikimate 5-2 dehydrogenase (aroe) from vibrio cholerae
|
| 14 | d1nvta1
|
|
 |
99.6 |
24 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 15 | c2rirA_
|
|
 |
99.5 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dipicolinate synthase, a chain;
PDBTitle: crystal structure of dipicolinate synthase, a chain, from bacillus2 subtilis
|
| 16 | c2i99A_
|
|
 |
99.5 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:mu-crystallin homolog;
PDBTitle: crystal structure of human mu_crystallin at 2.6 angstrom
|
| 17 | d1l7da1
|
|
 |
99.5 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 18 | c3d4oA_
|
|
 |
99.5 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dipicolinate synthase subunit a;
PDBTitle: crystal structure of dipicolinate synthase subunit a (np_243269.1)2 from bacillus halodurans at 2.10 a resolution
|
| 19 | d1nyta1
|
|
 |
99.4 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 20 | d1npya1
|
|
 |
99.4 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 21 | d1vi2a1 |
|
not modelled |
99.4 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 22 | d1pjca1 |
|
not modelled |
99.4 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 23 | d1p77a1 |
|
not modelled |
99.4 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 24 | d1x7da_ |
|
not modelled |
99.4 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Ornithine cyclodeaminase-like |
| 25 | c3pwzA_ |
|
not modelled |
99.3 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:shikimate dehydrogenase 3;
PDBTitle: crystal structure of an ael1 enzyme from pseudomonas putida
|
| 26 | c2eezG_ |
|
not modelled |
99.3 |
21 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:alanine dehydrogenase;
PDBTitle: crystal structure of alanine dehydrogenase from themus thermophilus
|
| 27 | c2hk8B_ |
|
not modelled |
99.3 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:shikimate dehydrogenase;
PDBTitle: crystal structure of shikimate dehydrogenase from aquifex2 aeolicus at 2.35 angstrom resolution
|
| 28 | d1omoa_ |
|
not modelled |
99.3 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Ornithine cyclodeaminase-like |
| 29 | c1nvtA_ |
|
not modelled |
99.3 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:shikimate 5'-dehydrogenase;
PDBTitle: crystal structure of shikimate dehydrogenase (aroe or2 mj1084) in complex with nadp+
|
| 30 | c3u62A_ |
|
not modelled |
99.2 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:shikimate dehydrogenase;
PDBTitle: crystal structure of shikimate dehydrogenase from thermotoga maritima
|
| 31 | c1p74B_ |
|
not modelled |
99.2 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:shikimate 5-dehydrogenase;
PDBTitle: crystal structure of shikimate dehydrogenase (aroe) from2 haemophilus influenzae
|
| 32 | c3tozA_ |
|
not modelled |
99.2 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:shikimate dehydrogenase;
PDBTitle: 2.2 angstrom crystal structure of shikimate 5-dehydrogenase from2 listeria monocytogenes in complex with nad.
|
| 33 | c1nytC_ |
|
not modelled |
99.2 |
22 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:shikimate 5-dehydrogenase;
PDBTitle: shikimate dehydrogenase aroe complexed with nadp+
|
| 34 | c3dfzB_ |
|
not modelled |
99.2 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:precorrin-2 dehydrogenase;
PDBTitle: sirc, precorrin-2 dehydrogenase
|
| 35 | c3hdjA_ |
|
not modelled |
99.1 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:probable ornithine cyclodeaminase;
PDBTitle: the crystal structure of probable ornithine cyclodeaminase from2 bordetella pertussis tohama i
|
| 36 | c1npyA_ |
|
not modelled |
99.1 |
23 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical shikimate 5-dehydrogenase-like
PDBTitle: structure of shikimate 5-dehydrogenase-like protein hi0607
|
| 37 | c2o7qA_ |
|
not modelled |
99.1 |
26 |
PDB header:oxidoreductase,transferase
Chain: A: PDB Molecule:bifunctional 3-dehydroquinate dehydratase/shikimate
PDBTitle: crystal structure of the a. thaliana dhq-dehydroshikimate-sdh-2 shikimate-nadp(h)
|
| 38 | c2ev9B_ |
|
not modelled |
99.1 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:shikimate 5-dehydrogenase;
PDBTitle: crystal structure of shikimate 5-dehydrogenase (aroe) from thermus2 thermophilus hb8 in complex with nadp(h) and shikimate
|
| 39 | c1vi2B_ |
|
not modelled |
99.1 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:shikimate 5-dehydrogenase 2;
PDBTitle: crystal structure of shikimate-5-dehydrogenase with nad
|
| 40 | c3p2yA_ |
|
not modelled |
99.1 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alanine dehydrogenase/pyridine nucleotide transhydrogenase;
PDBTitle: crystal structure of alanine dehydrogenase/pyridine nucleotide2 transhydrogenase from mycobacterium smegmatis
|
| 41 | c1d4fD_ |
|
not modelled |
99.0 |
16 |
PDB header:hydrolase
Chain: D: PDB Molecule:s-adenosylhomocysteine hydrolase;
PDBTitle: crystal structure of recombinant rat-liver d244e mutant s-2 adenosylhomocysteine hydrolase
|
| 42 | c4a26B_ |
|
not modelled |
99.0 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative c-1-tetrahydrofolate synthase, cytoplasmic;
PDBTitle: the crystal structure of leishmania major n5,n10-2 methylenetetrahydrofolate dehydrogenase/cyclohydrolase
|
| 43 | c2bruB_ |
|
not modelled |
99.0 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nad(p) transhydrogenase subunit alpha;
PDBTitle: complex of the domain i and domain iii of escherichia coli2 transhydrogenase
|
| 44 | c3fbtB_ |
|
not modelled |
98.9 |
16 |
PDB header:oxidoreductase, lyase
Chain: B: PDB Molecule:chorismate mutase and shikimate 5-dehydrogenase
PDBTitle: crystal structure of a chorismate mutase/shikimate 5-2 dehydrogenase fusion protein from clostridium3 acetobutylicum
|
| 45 | d1li4a1 |
|
not modelled |
98.9 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 46 | d1a4ia1 |
|
not modelled |
98.9 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 47 | c1pjcA_ |
|
not modelled |
98.9 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein (l-alanine dehydrogenase);
PDBTitle: l-alanine dehydrogenase complexed with nad
|
| 48 | d1leha1 |
|
not modelled |
98.9 |
12 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 49 | c2axqA_ |
|
not modelled |
98.9 |
28 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:saccharopine dehydrogenase;
PDBTitle: apo histidine-tagged saccharopine dehydrogenase (l-glu2 forming) from saccharomyces cerevisiae
|
| 50 | c1l7eC_ |
|
not modelled |
98.8 |
16 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:nicotinamide nucleotide transhydrogenase,
PDBTitle: crystal structure of r. rubrum transhydrogenase domain i2 with bound nadh
|
| 51 | c3n58D_ |
|
not modelled |
98.8 |
16 |
PDB header:hydrolase
Chain: D: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structure of s-adenosyl-l-homocysteine hydrolase from brucella2 melitensis in ternary complex with nad and adenosine, orthorhombic3 form
|
| 52 | c3l07B_ |
|
not modelled |
98.8 |
22 |
PDB header:oxidoreductase,hydrolase
Chain: B: PDB Molecule:bifunctional protein fold;
PDBTitle: methylenetetrahydrofolate dehydrogenase/methenyltetrahydrofolate2 cyclohydrolase, putative bifunctional protein fold from francisella3 tularensis.
|
| 53 | d1b0aa1 |
|
not modelled |
98.8 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 54 | c1v8bA_ |
|
not modelled |
98.8 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structure of a hydrolase
|
| 55 | c1e5lA_ |
|
not modelled |
98.8 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:saccharopine reductase;
PDBTitle: apo saccharopine reductase from magnaporthe grisea
|
| 56 | c3dhyC_ |
|
not modelled |
98.7 |
16 |
PDB header:hydrolase
Chain: C: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structures of mycobacterium tuberculosis s-adenosyl-l-2 homocysteine hydrolase in ternary complex with substrate and3 inhibitors
|
| 57 | c3oneA_ |
|
not modelled |
98.7 |
20 |
PDB header:hydrolase/hydrolase substrate
Chain: A: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structure of lupinus luteus s-adenosyl-l-homocysteine2 hydrolase in complex with adenine
|
| 58 | c1np3B_ |
|
not modelled |
98.7 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:ketol-acid reductoisomerase;
PDBTitle: crystal structure of class i acetohydroxy acid isomeroreductase from2 pseudomonas aeruginosa
|
| 59 | c3triB_ |
|
not modelled |
98.7 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:pyrroline-5-carboxylate reductase;
PDBTitle: structure of a pyrroline-5-carboxylate reductase (proc) from coxiella2 burnetii
|
| 60 | d1v8ba1 |
|
not modelled |
98.7 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 61 | c3d64A_ |
|
not modelled |
98.7 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structure of s-adenosyl-l-homocysteine hydrolase from2 burkholderia pseudomallei
|
| 62 | c3d1lB_ |
|
not modelled |
98.6 |
26 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative nadp oxidoreductase bf3122;
PDBTitle: crystal structure of putative nadp oxidoreductase bf3122 from2 bacteroides fragilis
|
| 63 | c4a5oB_ |
|
not modelled |
98.6 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:bifunctional protein fold;
PDBTitle: crystal structure of pseudomonas aeruginosa n5, n10-2 methylenetetrahydrofolate dehydrogenase-cyclohydrolase (fold)
|
| 64 | c1a4iB_ |
|
not modelled |
98.6 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:methylenetetrahydrofolate dehydrogenase /
PDBTitle: human tetrahydrofolate dehydrogenase / cyclohydrolase
|
| 65 | d1pjqa1 |
|
not modelled |
98.6 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Siroheme synthase N-terminal domain-like |
| 66 | c3b1fA_ |
|
not modelled |
98.6 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative prephenate dehydrogenase;
PDBTitle: crystal structure of prephenate dehydrogenase from streptococcus2 mutans
|
| 67 | c3cumA_ |
|
not modelled |
98.6 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable 3-hydroxyisobutyrate dehydrogenase;
PDBTitle: crystal structure of a possible 3-hydroxyisobutyrate dehydrogenase2 from pseudomonas aeruginosa pao1
|
| 68 | c3gt0A_ |
|
not modelled |
98.6 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pyrroline-5-carboxylate reductase;
PDBTitle: crystal structure of pyrroline 5-carboxylate reductase from bacillus2 cereus. northeast structural genomics consortium target bcr38b
|
| 69 | d1e5qa1 |
|
not modelled |
98.6 |
26 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 70 | c3p2oB_ |
|
not modelled |
98.6 |
21 |
PDB header:oxidoreductase, hydrolase
Chain: B: PDB Molecule:bifunctional protein fold;
PDBTitle: crystal structure of fold bifunctional protein from campylobacter2 jejuni
|
| 71 | d1luaa1 |
|
not modelled |
98.6 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 72 | c2f1kD_ |
|
not modelled |
98.6 |
21 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:prephenate dehydrogenase;
PDBTitle: crystal structure of synechocystis arogenate dehydrogenase
|
| 73 | c2g5cD_ |
|
not modelled |
98.6 |
17 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:prephenate dehydrogenase;
PDBTitle: crystal structure of prephenate dehydrogenase from aquifex aeolicus
|
| 74 | c3p2oA_ |
|
not modelled |
98.6 |
21 |
PDB header:oxidoreductase, hydrolase
Chain: A: PDB Molecule:bifunctional protein fold;
PDBTitle: crystal structure of fold bifunctional protein from campylobacter2 jejuni
|
| 75 | c2ag8A_ |
|
not modelled |
98.5 |
30 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pyrroline-5-carboxylate reductase;
PDBTitle: nadp complex of pyrroline-5-carboxylate reductase from neisseria2 meningitidis
|
| 76 | d1gdha1 |
|
not modelled |
98.5 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 77 | d1mx3a1 |
|
not modelled |
98.5 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 78 | c2g76A_ |
|
not modelled |
98.5 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-3-phosphoglycerate dehydrogenase;
PDBTitle: crystal structure of human 3-phosphoglycerate dehydrogenase
|
| 79 | d1ygya1 |
|
not modelled |
98.5 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 80 | c2ahrB_ |
|
not modelled |
98.5 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative pyrroline carboxylate reductase;
PDBTitle: crystal structures of 1-pyrroline-5-carboxylate reductase from human2 pathogen streptococcus pyogenes
|
| 81 | c3dttA_ |
|
not modelled |
98.5 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadp oxidoreductase;
PDBTitle: crystal structure of a putative f420 dependent nadp-reductase2 (arth_0613) from arthrobacter sp. fb24 at 1.70 a resolution
|
| 82 | d1c1da1 |
|
not modelled |
98.5 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 83 | d2naca1 |
|
not modelled |
98.5 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 84 | c2c2xB_ |
|
not modelled |
98.5 |
26 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:methylenetetrahydrofolate dehydrogenase-
PDBTitle: three dimensional structure of bifunctional2 methylenetetrahydrofolate dehydrogenase-cyclohydrolase3 from mycobacterium tuberculosis
|
| 85 | c1vpdA_ |
|
not modelled |
98.5 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:tartronate semialdehyde reductase;
PDBTitle: x-ray crystal structure of tartronate semialdehyde reductase2 [salmonella typhimurium lt2]
|
| 86 | d1uxja1 |
|
not modelled |
98.5 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 87 | c3dzbA_ |
|
not modelled |
98.5 |
18 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:prephenate dehydrogenase;
PDBTitle: crystal structure of prephenate dehydrogenase from streptococcus2 thermophilus
|
| 88 | c2eklA_ |
|
not modelled |
98.5 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-3-phosphoglycerate dehydrogenase;
PDBTitle: structure of st1218 protein from sulfolobus tokodaii
|
| 89 | c3fwnB_ |
|
not modelled |
98.5 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:6-phosphogluconate dehydrogenase, decarboxylating;
PDBTitle: dimeric 6-phosphogluconate dehydrogenase complexed with 6-2 phosphogluconate and 2'-monophosphoadenosine-5'-diphosphate
|
| 90 | d1np3a2 |
|
not modelled |
98.5 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 91 | c1b0aA_ |
|
not modelled |
98.5 |
14 |
PDB header:oxidoreductase,hydrolase
Chain: A: PDB Molecule:protein (fold bifunctional protein);
PDBTitle: 5,10, methylene-tetrahydropholate2 dehydrogenase/cyclohydrolase from e coli.
|
| 92 | c3g0oA_ |
|
not modelled |
98.4 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-hydroxyisobutyrate dehydrogenase;
PDBTitle: crystal structure of 3-hydroxyisobutyrate dehydrogenase2 (ygbj) from salmonella typhimurium
|
| 93 | c3l6dB_ |
|
not modelled |
98.4 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of putative oxidoreductase from pseudomonas putida2 kt2440
|
| 94 | d1bg6a2 |
|
not modelled |
98.4 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 95 | c1gdhA_ |
|
not modelled |
98.4 |
16 |
PDB header:oxidoreductase(choh (d)-nad(p)+ (a))
Chain: A: PDB Molecule:d-glycerate dehydrogenase;
PDBTitle: crystal structure of a nad-dependent d-glycerate2 dehydrogenase at 2.4 angstroms resolution
|
| 96 | d1jaya_ |
|
not modelled |
98.4 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 97 | c1wwkA_ |
|
not modelled |
98.4 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:phosphoglycerate dehydrogenase;
PDBTitle: crystal structure of phosphoglycerate dehydrogenase from pyrococcus2 horikoshii ot3
|
| 98 | c2iz1C_ |
|
not modelled |
98.4 |
15 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:6-phosphogluconate dehydrogenase, decarboxylating;
PDBTitle: 6pdh complexed with pex inhibitor synchrotron data
|
| 99 | c2uyyD_ |
|
not modelled |
98.4 |
16 |
PDB header:cytokine
Chain: D: PDB Molecule:n-pac protein;
PDBTitle: structure of the cytokine-like nuclear factor n-pac
|
| 100 | c2vhyB_ |
|
not modelled |
98.4 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:alanine dehydrogenase;
PDBTitle: crystal structure of apo l-alanine dehydrogenase from2 mycobacterium tuberculosis
|
| 101 | d1yqga2 |
|
not modelled |
98.4 |
28 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 102 | c3ggpA_ |
|
not modelled |
98.4 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:prephenate dehydrogenase;
PDBTitle: crystal structure of prephenate dehydrogenase from a. aeolicus in2 complex with hydroxyphenyl propionate and nad+
|
| 103 | c1bxgA_ |
|
not modelled |
98.4 |
15 |
PDB header:amino acid dehydrogenase
Chain: A: PDB Molecule:phenylalanine dehydrogenase;
PDBTitle: phenylalanine dehydrogenase structure in ternary complex2 with nad+ and beta-phenylpropionate
|
| 104 | d2pgda2 |
|
not modelled |
98.4 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 105 | c3n7uD_ |
|
not modelled |
98.4 |
20 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:formate dehydrogenase;
PDBTitle: nad-dependent formate dehydrogenase from higher-plant arabidopsis2 thaliana in complex with nad and azide
|
| 106 | d3cuma2 |
|
not modelled |
98.4 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 107 | c2d0iC_ |
|
not modelled |
98.4 |
23 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:dehydrogenase;
PDBTitle: crystal structure ph0520 protein from pyrococcus horikoshii ot3
|
| 108 | d1j4aa1 |
|
not modelled |
98.4 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 109 | c1pgjA_ |
|
not modelled |
98.4 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:6-phosphogluconate dehydrogenase;
PDBTitle: x-ray structure of 6-phosphogluconate dehydrogenase from the protozoan2 parasite t. brucei
|
| 110 | c2cvzD_ |
|
not modelled |
98.4 |
27 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:3-hydroxyisobutyrate dehydrogenase;
PDBTitle: structure of hydroxyisobutyrate dehydrogenase from thermus2 thermophilus hb8
|
| 111 | c1bg6A_ |
|
not modelled |
98.4 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:n-(1-d-carboxylethyl)-l-norvaline dehydrogenase;
PDBTitle: crystal structure of the n-(1-d-carboxylethyl)-l-norvaline2 dehydrogenase from arthrobacter sp. strain 1c
|
| 112 | c3ktdC_ |
|
not modelled |
98.4 |
16 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:prephenate dehydrogenase;
PDBTitle: crystal structure of a putative prephenate dehydrogenase (cgl0226)2 from corynebacterium glutamicum atcc 13032 at 2.60 a resolution
|
| 113 | c1edzA_ |
|
not modelled |
98.4 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:5,10-methylenetetrahydrofolate dehydrogenase;
PDBTitle: structure of the nad-dependent 5,10-2 methylenetetrahydrofolate dehydrogenase from saccharomyces3 cerevisiae
|
| 114 | c2dbqA_ |
|
not modelled |
98.4 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glyoxylate reductase;
PDBTitle: crystal structure of glyoxylate reductase (ph0597) from pyrococcus2 horikoshii ot3, complexed with nadp (i41)
|
| 115 | d2dlda1 |
|
not modelled |
98.4 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 116 | c2j6iC_ |
|
not modelled |
98.4 |
22 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:formate dehydrogenase;
PDBTitle: candida boidinii formate dehydrogenase (fdh) c-terminal2 mutant
|
| 117 | c3c24A_ |
|
not modelled |
98.4 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of a putative oxidoreductase (yp_511008.1) from2 jannaschia sp. ccs1 at 1.62 a resolution
|
| 118 | d1dxya1 |
|
not modelled |
98.3 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 119 | d2ahra2 |
|
not modelled |
98.3 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 120 | c3gg9C_ |
|
not modelled |
98.3 |
23 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:d-3-phosphoglycerate dehydrogenase oxidoreductase protein;
PDBTitle: crystal structure of putative d-3-phosphoglycerate dehydrogenase2 oxidoreductase from ralstonia solanacearum
|