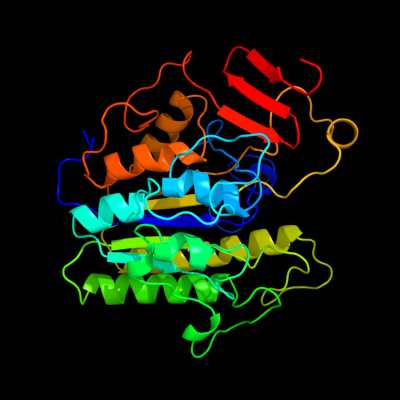
1 c3lxqB_
99.1
13
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein vp1736;PDBTitle: the crystal structure of a protein in the alkaline2 phosphatase superfamily from vibrio parahaemolyticus to3 1.95a
2 c2w8dB_
98.9
15
PDB header: transferaseChain: B: PDB Molecule: processed glycerol phosphate lipoteichoic acid synthase 2;PDBTitle: distinct and essential morphogenic functions for wall- and2 lipo-teichoic acids in bacillus subtilis
3 c2w5tA_
98.6
14
PDB header: transferaseChain: A: PDB Molecule: processed glycerol phosphate lipoteichoic acidPDBTitle: structure-based mechanism of lipoteichoic acid synthesis by2 staphylococcus aureus ltas.
4 c3ed4A_
98.2
15
PDB header: transferaseChain: A: PDB Molecule: arylsulfatase;PDBTitle: crystal structure of putative arylsulfatase from escherichia coli
5 c2qzuA_
98.2
14
PDB header: hydrolaseChain: A: PDB Molecule: putative sulfatase yidj;PDBTitle: crystal structure of the putative sulfatase yidj from bacteroides2 fragilis. northeast structural genomics consortium target bfr123
6 d1auka_
98.0
15
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Arylsulfatase7 d1fsua_
97.8
15
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Arylsulfatase8 c2vqrA_
97.5
20
PDB header: hydrolaseChain: A: PDB Molecule: putative sulfatase;PDBTitle: crystal structure of a phosphonate monoester hydrolase2 from rhizobium leguminosarum: a new member of the3 alkaline phosphatase superfamily
9 d1hdha_
97.4
18
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Arylsulfatase10 c3b5qB_
97.1
18
PDB header: hydrolaseChain: B: PDB Molecule: putative sulfatase yidj;PDBTitle: crystal structure of a putative sulfatase (np_810509.1)2 from bacteroides thetaiotaomicron vpi-5482 at 2.40 a3 resolution
11 d1o98a2
96.1
13
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: 2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, catalytic domain12 c3q3qA_
94.8
21
PDB header: hydrolaseChain: A: PDB Molecule: alkaline phosphatase;PDBTitle: crystal structure of spap: an novel alkaline phosphatase from2 bacterium sphingomonas sp. strain bsar-1
13 d1p49a_
92.9
13
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Arylsulfatase14 c2xrgA_
92.6
14
PDB header: hydrolaseChain: A: PDB Molecule: ectonucleotide pyrophosphatase/phosphodiesterase familyPDBTitle: crystal structure of autotaxin (enpp2) in complex with the2 ha155 boronic acid inhibitor
15 c3szzA_
91.8
15
PDB header: hydrolaseChain: A: PDB Molecule: phosphonoacetate hydrolase;PDBTitle: crystal structure of phosphonoacetate hydrolase from sinorhizobium2 meliloti 1021 in complex with acetate
16 d1ei6a_
90.7
15
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Phosphonoacetate hydrolase17 d2i09a1
90.1
11
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: DeoB catalytic domain-like18 c2gsoB_
90.0
10
PDB header: hydrolaseChain: B: PDB Molecule: phosphodiesterase-nucleotide pyrophosphatase;PDBTitle: structure of xac nucleotide2 pyrophosphatase/phosphodiesterase in complex with vanadate
19 c2i09A_
71.9
13
PDB header: isomeraseChain: A: PDB Molecule: phosphopentomutase;PDBTitle: crystal structure of putative phosphopentomutase from streptococcus2 mutans
20 d1esfa1
71.3
26
Fold: OB-foldSuperfamily: Bacterial enterotoxinsFamily: Superantigen toxins, N-terminal domain21 c2xr9A_
not modelled
69.3
16
PDB header: hydrolaseChain: A: PDB Molecule: ectonucleotide pyrophosphatase/phosphodiesterase familyPDBTitle: crystal structure of autotaxin (enpp2)
22 d1qhfa_
not modelled
68.4
25
Fold: Phosphoglycerate mutase-likeSuperfamily: Phosphoglycerate mutase-likeFamily: Cofactor-dependent phosphoglycerate mutase23 c3f3kA_
not modelled
67.9
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein ykr043c;PDBTitle: the structure of uncharacterized protein ykr043c from saccharomyces2 cerevisiae.
24 c3c7tB_
not modelled
56.6
21
PDB header: hydrolaseChain: B: PDB Molecule: ecdysteroid-phosphate phosphatase;PDBTitle: crystal structure of the ecdysone phosphate phosphatase, eppase, from2 bombix mori in complex with tungstate
25 c1yjxD_
not modelled
52.5
28
PDB header: isomerase, hydrolaseChain: D: PDB Molecule: phosphoglycerate mutase 1;PDBTitle: crystal structure of human b type phosphoglycerate mutase
26 d2hhja1
not modelled
43.8
18
Fold: Phosphoglycerate mutase-likeSuperfamily: Phosphoglycerate mutase-likeFamily: Cofactor-dependent phosphoglycerate mutase27 d1klud1
not modelled
43.4
24
Fold: OB-foldSuperfamily: Bacterial enterotoxinsFamily: Superantigen toxins, N-terminal domain28 d2aq2b1
not modelled
42.2
24
Fold: OB-foldSuperfamily: Bacterial enterotoxinsFamily: Superantigen toxins, N-terminal domain29 d1ck1a1
not modelled
41.6
25
Fold: OB-foldSuperfamily: Bacterial enterotoxinsFamily: Superantigen toxins, N-terminal domain30 d1xq9a_
not modelled
40.4
23
Fold: Phosphoglycerate mutase-likeSuperfamily: Phosphoglycerate mutase-likeFamily: Cofactor-dependent phosphoglycerate mutase31 c2ikqA_
not modelled
38.4
21
PDB header: signaling protein, immune systemChain: A: PDB Molecule: suppressor of t-cell receptor signaling 1;PDBTitle: crystal structure of mouse sts-1 pgm domain in complex with phosphate
32 c3d4iD_
not modelled
35.4
19
PDB header: hydrolaseChain: D: PDB Molecule: sts-2 protein;PDBTitle: crystal structure of the 2h-phosphatase domain of sts-2
33 d1s0yb_
not modelled
30.3
26
Fold: Tautomerase/MIFSuperfamily: Tautomerase/MIFFamily: 4-oxalocrotonate tautomerase-like34 c3ej9D_
not modelled
29.5
26
PDB header: hydrolaseChain: D: PDB Molecule: beta-subunit of trans-3-chloroacrylic acid dehalogenase;PDBTitle: structural and mechanistic analysis of trans-3-chloroacrylic acid2 dehalogenase activity
35 d1riia_
not modelled
28.7
23
Fold: Phosphoglycerate mutase-likeSuperfamily: Phosphoglycerate mutase-likeFamily: Cofactor-dependent phosphoglycerate mutase36 d1ugpb_
not modelled
27.3
9
Fold: SH3-like barrelSuperfamily: Electron transport accessory proteinsFamily: Nitrile hydratase beta chain37 c3czpA_
not modelled
27.2
13
PDB header: transferaseChain: A: PDB Molecule: putative polyphosphate kinase 2;PDBTitle: crystal structure of putative polyphosphate kinase 2 from pseudomonas2 aeruginosa pa01
38 d1bifa2
not modelled
26.8
24
Fold: Phosphoglycerate mutase-likeSuperfamily: Phosphoglycerate mutase-likeFamily: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase, phosphatase domain39 c1bplA_
not modelled
24.6
27
PDB header: glycosyltransferaseChain: A: PDB Molecule: alpha-1,4-glucan-4-glucanohydrolase;PDBTitle: glycosyltransferase
40 d2bvya2
not modelled
24.2
19
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases41 d1xbta1
not modelled
23.6
32
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Type II thymidine kinase42 d1e58a_
not modelled
23.5
20
Fold: Phosphoglycerate mutase-likeSuperfamily: Phosphoglycerate mutase-likeFamily: Cofactor-dependent phosphoglycerate mutase43 c3sr2A_
not modelled
23.2
23
PDB header: dna binding protein/protein bindingChain: A: PDB Molecule: dna repair protein xrcc4;PDBTitle: crystal structure of human xlf-xrcc4 complex
44 c2bvtB_
not modelled
23.1
17
PDB header: hydrolaseChain: B: PDB Molecule: beta-1,4-mannanase;PDBTitle: the structure of a modular endo-beta-1,4-mannanase from2 cellulomonas fimi explains the product specificity of3 glycoside hydrolase family 26 mannanases.
45 d1ud2a2
not modelled
22.5
23
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain46 d1bxta1
not modelled
22.3
30
Fold: OB-foldSuperfamily: Bacterial enterotoxinsFamily: Superantigen toxins, N-terminal domain47 c3eznB_
not modelled
22.3
22
PDB header: isomeraseChain: B: PDB Molecule: 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase;PDBTitle: crystal structure of phosphoglyceromutase from burkholderia2 pseudomallei 1710b
48 c3mudA_
not modelled
21.8
23
PDB header: contractile proteinChain: A: PDB Molecule: dna repair protein xrcc4, tropomyosin alpha-1 chain;PDBTitle: structure of the tropomyosin overlap complex from chicken smooth2 muscle
49 c3k8kB_
not modelled
21.6
27
PDB header: membrane proteinChain: B: PDB Molecule: alpha-amylase, susg;PDBTitle: crystal structure of susg
50 c1cygA_
not modelled
21.1
19
PDB header: glycosyltransferaseChain: A: PDB Molecule: cyclodextrin glucanotransferase;PDBTitle: cyclodextrin glucanotransferase (e.c.2.4.1.19) (cgtase)
51 c3ll4B_
not modelled
20.7
19
PDB header: hydrolaseChain: B: PDB Molecule: uncharacterized protein ykr043c;PDBTitle: structure of the h13a mutant of ykr043c in complex with fructose-1,6-2 bisphosphate
52 c1jd7A_
not modelled
18.8
12
PDB header: hydrolaseChain: A: PDB Molecule: alpha-amylase;PDBTitle: crystal structure analysis of the mutant k300r of2 pseudoalteromonas haloplanctis alpha-amylase
53 c1k6mA_
not modelled
18.6
24
PDB header: transferase, hydrolaseChain: A: PDB Molecule: 6-phosphofructo-2-kinase/fructose-2,6-PDBTitle: crystal structure of human liver 6-phosphofructo-2-2 kinase/fructose-2,6-bisphosphatase
54 c1tcmB_
not modelled
18.3
15
PDB header: glycosyltransferaseChain: B: PDB Molecule: cyclodextrin glycosyltransferase;PDBTitle: cyclodextrin glycosyltransferase w616a mutant from bacillus2 circulans strain 251
55 d1mxga2
not modelled
18.2
28
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain56 d1gjwa2
not modelled
17.9
23
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain57 d1ob0a2
not modelled
17.6
27
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain58 c3qz9D_
not modelled
17.4
7
PDB header: lyaseChain: D: PDB Molecule: co-type nitrile hydratase beta subunit;PDBTitle: crystal structure of co-type nitrile hydratase beta-y215f from2 pseudomonas putida.
59 c1mwoA_
not modelled
17.4
31
PDB header: hydrolaseChain: A: PDB Molecule: alpha amylase;PDBTitle: crystal structure analysis of the hyperthermostable2 pyrocoocus woesei alpha-amylase
60 c2wskA_
not modelled
17.2
29
PDB header: hydrolaseChain: A: PDB Molecule: glycogen debranching enzyme;PDBTitle: crystal structure of glycogen debranching enzyme glgx from2 escherichia coli k-12
61 c3rgiA_
not modelled
16.3
15
PDB header: transferaseChain: A: PDB Molecule: disd protein;PDBTitle: trans-acting transferase from disorazole synthase
62 c1jaeA_
not modelled
16.1
15
PDB header: glycosidaseChain: A: PDB Molecule: alpha-amylase;PDBTitle: structure of tenebrio molitor larval alpha-amylase
63 c1qhoA_
not modelled
16.0
31
PDB header: hydrolaseChain: A: PDB Molecule: alpha-amylase;PDBTitle: five-domain alpha-amylase from bacillus stearothermophilus,2 maltose/acarbose complex
64 c2qpuB_
not modelled
15.8
24
PDB header: hydrolaseChain: B: PDB Molecule: alpha-amylase type a isozyme;PDBTitle: sugar tongs mutant s378p in complex with acarbose
65 d2fhfa5
not modelled
15.8
21
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain66 c3k25B_
not modelled
15.7
29
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: slr1438 protein;PDBTitle: crystal structure of slr1438 protein from synechocystis sp. pcc 6803,2 northeast structural genomics consortium target sgr112
67 c3d8hB_
not modelled
15.5
19
PDB header: isomeraseChain: B: PDB Molecule: glycolytic phosphoglycerate mutase;PDBTitle: crystal structure of phosphoglycerate mutase from cryptosporidium2 parvum, cgd7_4270
68 d1avaa2
not modelled
15.4
23
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain69 d3pgma_
not modelled
15.1
24
Fold: Phosphoglycerate mutase-likeSuperfamily: Phosphoglycerate mutase-likeFamily: Cofactor-dependent phosphoglycerate mutase70 c1jdaA_
not modelled
14.9
35
PDB header: hydrolaseChain: A: PDB Molecule: 1,4-alpha maltotetrahydrolase;PDBTitle: maltotetraose-forming exo-amylase
71 d1hvxa2
not modelled
14.6
19
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain72 d1q6za1
not modelled
14.6
20
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain73 c1bf2A_
not modelled
14.4
31
PDB header: hydrolaseChain: A: PDB Molecule: isoamylase;PDBTitle: structure of pseudomonas isoamylase
74 d1cyga4
not modelled
14.2
17
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain75 c3bmwA_
not modelled
13.9
19
PDB header: transferaseChain: A: PDB Molecule: cyclomaltodextrin glucanotransferase;PDBTitle: cyclodextrin glycosyl transferase from thermoanerobacterium2 thermosulfurigenes em1 mutant s77p complexed with a maltoheptaose3 inhibitor
76 c2ya0A_
not modelled
13.9
25
PDB header: hydrolaseChain: A: PDB Molecule: putative alkaline amylopullulanase;PDBTitle: catalytic module of the multi-modular glycogen-degrading2 pneumococcal virulence factor spua
77 c3bv6D_
not modelled
13.7
14
PDB header: hydrolaseChain: D: PDB Molecule: metal-dependent hydrolase;PDBTitle: crystal structure of uncharacterized metallo protein from vibrio2 cholerae with beta-lactamase like fold
78 c2i1vB_
not modelled
13.6
32
PDB header: transferase, hydrolaseChain: B: PDB Molecule: 6-phosphofructo-2-kinase/fructose-2,6-PDBTitle: crystal structure of pfkfb3 in complex with adp and2 fructose-2,6-bisphosphate
79 c2aaaA_
not modelled
13.6
38
PDB header: glycosidaseChain: A: PDB Molecule: alpha-amylase;PDBTitle: calcium binding in alpha-amylases: an x-ray diffraction2 study at 2.1 angstroms resolution of two enzymes from3 aspergillus
80 d2gjpa2
not modelled
13.5
27
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain81 c3faxA_
not modelled
13.5
31
PDB header: hydrolaseChain: A: PDB Molecule: reticulocyte binding protein;PDBTitle: the crystal structure of gbs pullulanase sap in complex with2 maltotetraose
82 d2btya1
not modelled
13.5
31
Fold: Carbamate kinase-likeSuperfamily: Carbamate kinase-likeFamily: N-acetyl-l-glutamate kinase83 c2wcsA_
not modelled
13.4
14
PDB header: hydrolaseChain: A: PDB Molecule: alpha amylase, catalytic region;PDBTitle: crystal structure of debranching enzyme from nostoc2 punctiforme (npde)
84 c3blpX_
not modelled
13.2
27
PDB header: hydrolaseChain: X: PDB Molecule: alpha-amylase 1;PDBTitle: role of aromatic residues in human salivary alpha-amylase
85 d3bmva4
not modelled
13.0
17
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain86 c3q4fG_
not modelled
12.9
23
PDB header: dna binding protein/protein bindingChain: G: PDB Molecule: dna repair protein xrcc4;PDBTitle: crystal structure of xrcc4/xlf-cernunnos complex
87 c2b78A_
not modelled
12.4
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein smu.776;PDBTitle: a putative sam-dependent methyltransferase from2 streptococcus mutans
88 d1iv8a2
not modelled
12.3
19
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain89 d2io8a1
not modelled
12.2
24
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: Glutathionylspermidine synthase substrate-binding domain-like90 d1e43a2
not modelled
12.0
27
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain91 d3bzka5
not modelled
11.8
37
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Tex RuvX-like domain-like92 c1w4rC_
not modelled
11.8
40
PDB header: transferaseChain: C: PDB Molecule: thymidine kinase;PDBTitle: structure of a type ii thymidine kinase with bound dttp
93 d2bhua3
not modelled
11.8
27
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain94 c3dhuC_
not modelled
11.7
24
PDB header: hydrolaseChain: C: PDB Molecule: alpha-amylase;PDBTitle: crystal structure of an alpha-amylase from lactobacillus2 plantarum
95 c3m07A_
not modelled
11.7
29
PDB header: unknown functionChain: A: PDB Molecule: putative alpha amylase;PDBTitle: 1.4 angstrom resolution crystal structure of putative alpha2 amylase from salmonella typhimurium.
96 d1k6ma2
not modelled
11.5
21
Fold: Phosphoglycerate mutase-likeSuperfamily: Phosphoglycerate mutase-likeFamily: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase, phosphatase domain97 d1m7xa3
not modelled
11.5
27
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain98 c1o7dC_
not modelled
11.5
23
PDB header: hydrolaseChain: C: PDB Molecule: lysosomal alpha-mannosidase;PDBTitle: the structure of the bovine lysosomal a-mannosidase2 suggests a novel mechanism for low ph activation
99 c3rhfB_
not modelled
11.4
25
PDB header: transferaseChain: B: PDB Molecule: putative polyphosphate kinase 2 family protein;PDBTitle: crystal structure of polyphosphate kinase 2 from arthrobacter2 aurescens tc1