| 1 | d1e19a_
|
|
 |
100.0 |
45 |
Fold:Carbamate kinase-like
Superfamily:Carbamate kinase-like
Family:Carbamate kinase |
| 2 | c2e9yA_
|
|
 |
100.0 |
38 |
PDB header:transferase
Chain: A: PDB Molecule:carbamate kinase;
PDBTitle: crystal structure of project ape1968 from aeropyrum pernix k1
|
| 3 | d1b7ba_
|
|
 |
100.0 |
44 |
Fold:Carbamate kinase-like
Superfamily:Carbamate kinase-like
Family:Carbamate kinase |
| 4 | c3kzfC_
|
|
 |
100.0 |
41 |
PDB header:transferase
Chain: C: PDB Molecule:carbamate kinase;
PDBTitle: structure of giardia carbamate kinase
|
| 5 | d2bufa1
|
|
 |
100.0 |
22 |
Fold:Carbamate kinase-like
Superfamily:Carbamate kinase-like
Family:N-acetyl-l-glutamate kinase |
| 6 | c2rd5A_
|
|
 |
100.0 |
20 |
PDB header:protein binding
Chain: A: PDB Molecule:acetylglutamate kinase-like protein;
PDBTitle: structural basis for the regulation of n-acetylglutamate kinase by pii2 in arabidopsis thaliana
|
| 7 | d2ap9a1
|
|
 |
100.0 |
20 |
Fold:Carbamate kinase-like
Superfamily:Carbamate kinase-like
Family:N-acetyl-l-glutamate kinase |
| 8 | c2r98A_
|
|
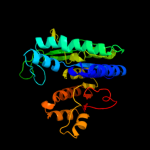 |
100.0 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:putative acetylglutamate synthase;
PDBTitle: crystal structure of n-acetylglutamate synthase (selenomet2 substituted) from neisseria gonorrhoeae
|
| 9 | d1gs5a_
|
|
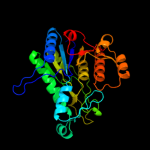 |
100.0 |
21 |
Fold:Carbamate kinase-like
Superfamily:Carbamate kinase-like
Family:N-acetyl-l-glutamate kinase |
| 10 | c2v5hB_
|
|
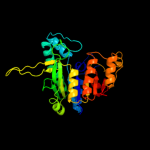 |
100.0 |
20 |
PDB header:transcription
Chain: B: PDB Molecule:acetylglutamate kinase;
PDBTitle: controlling the storage of nitrogen as arginine: the2 complex of pii and acetylglutamate kinase from3 synechococcus elongatus pcc 7942
|
| 11 | d2hmfa1
|
|
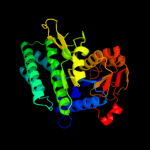 |
100.0 |
16 |
Fold:Carbamate kinase-like
Superfamily:Carbamate kinase-like
Family:PyrH-like |
| 12 | d2btya1
|
|
 |
100.0 |
18 |
Fold:Carbamate kinase-like
Superfamily:Carbamate kinase-like
Family:N-acetyl-l-glutamate kinase |
| 13 | c3l86A_
|
|
 |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:acetylglutamate kinase;
PDBTitle: the crystal structure of smu.665 from streptococcus mutans ua159
|
| 14 | c2j5tF_
|
|
 |
100.0 |
18 |
PDB header:transferase
Chain: F: PDB Molecule:glutamate 5-kinase;
PDBTitle: glutamate 5-kinase from escherichia coli complexed with2 glutamate
|
| 15 | c3c1nA_
|
|
 |
100.0 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:probable aspartokinase;
PDBTitle: crystal structure of allosteric inhibition threonine-sensitive2 aspartokinase from methanococcus jannaschii with l-threonine
|
| 16 | c2egxA_
|
|
 |
100.0 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative acetylglutamate kinase;
PDBTitle: crystal structure of the putative acetylglutamate kinase from thermus2 thermophilus
|
| 17 | c2w21A_
|
|
 |
100.0 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:glutamate 5-kinase;
PDBTitle: crystal structure of the aminoacid kinase domain of the2 glutamate 5 kinase of escherichia coli.
|
| 18 | d2cdqa1
|
|
 |
100.0 |
14 |
Fold:Carbamate kinase-like
Superfamily:Carbamate kinase-like
Family:PyrH-like |
| 19 | d2j0wa1
|
|
 |
100.0 |
18 |
Fold:Carbamate kinase-like
Superfamily:Carbamate kinase-like
Family:PyrH-like |
| 20 | c3l76B_
|
|
 |
100.0 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:aspartokinase;
PDBTitle: crystal structure of aspartate kinase from synechocystis
|
| 21 | d2bnea1 |
|
not modelled |
100.0 |
15 |
Fold:Carbamate kinase-like
Superfamily:Carbamate kinase-like
Family:PyrH-like |
| 22 | c2cdqB_ |
|
not modelled |
100.0 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:aspartokinase;
PDBTitle: crystal structure of arabidopsis thaliana aspartate kinase2 complexed with lysine and s-adenosylmethionine
|
| 23 | c2j0wA_ |
|
not modelled |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:lysine-sensitive aspartokinase 3;
PDBTitle: crystal structure of e. coli aspartokinase iii in complex2 with aspartate and adp (r-state)
|
| 24 | d1ybda1 |
|
not modelled |
100.0 |
16 |
Fold:Carbamate kinase-like
Superfamily:Carbamate kinase-like
Family:PyrH-like |
| 25 | c3ab4K_ |
|
not modelled |
100.0 |
19 |
PDB header:transferase
Chain: K: PDB Molecule:aspartokinase;
PDBTitle: crystal structure of feedback inhibition resistant mutant of aspartate2 kinase from corynebacterium glutamicum in complex with lysine and3 threonine
|
| 26 | d1z9da1 |
|
not modelled |
100.0 |
17 |
Fold:Carbamate kinase-like
Superfamily:Carbamate kinase-like
Family:PyrH-like |
| 27 | c3ll5C_ |
|
not modelled |
100.0 |
18 |
PDB header:transferase
Chain: C: PDB Molecule:gamma-glutamyl kinase related protein;
PDBTitle: crystal structure of t. acidophilum isopentenyl phosphate kinase2 product complex
|
| 28 | c3ll9A_ |
|
not modelled |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:isopentenyl phosphate kinase;
PDBTitle: x-ray structures of isopentenyl phosphate kinase
|
| 29 | c3ek5A_ |
|
not modelled |
100.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:uridylate kinase;
PDBTitle: unique gtp-binding pocket and allostery of ump kinase from a gram-2 negative phytopathogen bacterium
|
| 30 | d2akoa1 |
|
not modelled |
100.0 |
18 |
Fold:Carbamate kinase-like
Superfamily:Carbamate kinase-like
Family:PyrH-like |
| 31 | c2jjxC_ |
|
not modelled |
100.0 |
18 |
PDB header:transferase
Chain: C: PDB Molecule:uridylate kinase;
PDBTitle: the crystal structure of ump kinase from bacillus anthracis2 (ba1797)
|
| 32 | c3k4yB_ |
|
not modelled |
100.0 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:isopentenyl phosphate kinase;
PDBTitle: crystal structure of isopentenyl phosphate kinase from m. jannaschii2 in complex with ipp
|
| 33 | c3nwyB_ |
|
not modelled |
100.0 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:uridylate kinase;
PDBTitle: structure and allosteric regulation of the uridine monophosphate2 kinase from mycobacterium tuberculosis
|
| 34 | d2a1fa1 |
|
not modelled |
100.0 |
18 |
Fold:Carbamate kinase-like
Superfamily:Carbamate kinase-like
Family:PyrH-like |
| 35 | c2ogxB_ |
|
not modelled |
100.0 |
19 |
PDB header:metal binding protein
Chain: B: PDB Molecule:molybdenum storage protein subunit beta;
PDBTitle: the crystal structure of the molybdenum storage protein from2 azotobacter vinelandii loaded with polyoxotungstates (wsto)
|
| 36 | c3d40A_ |
|
not modelled |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:foma protein;
PDBTitle: crystal structure of fosfomycin resistance kinase foma from2 streptomyces wedmorensis complexed with diphosphate
|
| 37 | d2ij9a1 |
|
not modelled |
100.0 |
25 |
Fold:Carbamate kinase-like
Superfamily:Carbamate kinase-like
Family:PyrH-like |
| 38 | d2brxa1 |
|
not modelled |
100.0 |
22 |
Fold:Carbamate kinase-like
Superfamily:Carbamate kinase-like
Family:PyrH-like |
| 39 | c2va1A_ |
|
not modelled |
100.0 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:uridylate kinase;
PDBTitle: crystal structure of ump kinase from ureaplasma parvum
|
| 40 | c2ogxA_ |
|
not modelled |
100.0 |
20 |
PDB header:metal binding protein
Chain: A: PDB Molecule:molybdenum storage protein subunit alpha;
PDBTitle: the crystal structure of the molybdenum storage protein from2 azotobacter vinelandii loaded with polyoxotungstates (wsto)
|
| 41 | c2j4kC_ |
|
not modelled |
100.0 |
19 |
PDB header:transferase
Chain: C: PDB Molecule:uridylate kinase;
PDBTitle: crystal structure of uridylate kinase from sulfolobus2 solfataricus in complex with ump to 2.2 angstrom3 resolution
|
| 42 | c3bq9A_ |
|
not modelled |
56.9 |
28 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:predicted rossmann fold nucleotide-binding domain-
PDBTitle: crystal structure of predicted nucleotide-binding protein from2 idiomarina baltica os145
|
| 43 | d2zdra2 |
|
not modelled |
52.9 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:NeuB-like |
| 44 | d1iowa1 |
|
not modelled |
44.3 |
20 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:D-Alanine ligase N-terminal domain |
| 45 | c2dlnA_ |
|
not modelled |
41.1 |
20 |
PDB header:ligase(peptidoglycan synthesis)
Chain: A: PDB Molecule:d-alanine--d-alanine ligase;
PDBTitle: vancomycin resistance: structure of d-alanine:d-alanine2 ligase at 2.3 angstroms resolution
|
| 46 | c4a1aI_ |
|
not modelled |
38.5 |
29 |
PDB header:ribosome
Chain: I: PDB Molecule:60s ribosomal protein l13a;
PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 3.
|
| 47 | d1xpja_ |
|
not modelled |
35.5 |
13 |
Fold:HAD-like
Superfamily:HAD-like
Family:Hypothetical protein VC0232 |
| 48 | c3mn1B_ |
|
not modelled |
33.4 |
12 |
PDB header:hydrolase
Chain: B: PDB Molecule:probable yrbi family phosphatase;
PDBTitle: crystal structure of probable yrbi family phosphatase from pseudomonas2 syringae pv.phaseolica 1448a
|
| 49 | c2re2A_ |
|
not modelled |
26.8 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:uncharacterized protein ta1041;
PDBTitle: crystal structure of a putative iron-molybdenum cofactor (femo-co)2 dinitrogenase (ta1041m) from thermoplasma acidophilum dsm 1728 at3 1.30 a resolution
|
| 50 | c3dnpA_ |
|
not modelled |
26.0 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:stress response protein yhax;
PDBTitle: crystal structure of stress response protein yhax from bacillus2 subtilis
|
| 51 | c2yx6C_ |
|
not modelled |
24.3 |
5 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:hypothetical protein ph0822;
PDBTitle: crystal structure of ph0822
|
| 52 | d2b0ja2 |
|
not modelled |
23.8 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 53 | c3r4cA_ |
|
not modelled |
21.8 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:hydrolase, haloacid dehalogenase-like hydrolase;
PDBTitle: divergence of structure and function among phosphatases of the2 haloalkanoate (had) enzyme superfamily: analysis of bt1666 from3 bacteroides thetaiotaomicron
|
| 54 | d1t3va_ |
|
not modelled |
21.5 |
9 |
Fold:Ribonuclease H-like motif
Superfamily:Nitrogenase accessory factor-like
Family:MTH1175-like |
| 55 | d1o13a_ |
|
not modelled |
21.3 |
9 |
Fold:Ribonuclease H-like motif
Superfamily:Nitrogenase accessory factor-like
Family:MTH1175-like |
| 56 | c3izcK_ |
|
not modelled |
21.0 |
26 |
PDB header:ribosome
Chain: K: PDB Molecule:60s ribosomal protein rpl16 (l13p);
PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
|
| 57 | d2nx2a1 |
|
not modelled |
20.6 |
20 |
Fold:MCP/YpsA-like
Superfamily:MCP/YpsA-like
Family:YpsA-like |
| 58 | c2q5cA_ |
|
not modelled |
19.0 |
11 |
PDB header:transcription
Chain: A: PDB Molecule:ntrc family transcriptional regulator;
PDBTitle: crystal structure of ntrc family transcriptional regulator from2 clostridium acetobutylicum
|
| 59 | c2q4dB_ |
|
not modelled |
18.9 |
19 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:lysine decarboxylase-like protein at5g11950;
PDBTitle: ensemble refinement of the crystal structure of a lysine2 decarboxylase-like protein from arabidopsis thaliana gene at5g11950
|
| 60 | c3f46A_ |
|
not modelled |
18.8 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:5,10-methenyltetrahydromethanopterin hydrogenase;
PDBTitle: the crystal structure of c176a mutated [fe]-hydrogenase (hmd)2 holoenzyme from methanocaldococcus jannaschii
|
| 61 | d1rkqa_ |
|
not modelled |
18.7 |
26 |
Fold:HAD-like
Superfamily:HAD-like
Family:Predicted hydrolases Cof |
| 62 | c2q4oA_ |
|
not modelled |
18.7 |
24 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein at2g37210/t2n18.3;
PDBTitle: ensemble refinement of the crystal structure of a lysine2 decarboxylase-like protein from arabidopsis thaliana gene at2g37210
|
| 63 | d2q4oa1 |
|
not modelled |
18.7 |
24 |
Fold:MCP/YpsA-like
Superfamily:MCP/YpsA-like
Family:MoCo carrier protein-like |
| 64 | c3fzqA_ |
|
not modelled |
18.3 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative hydrolase;
PDBTitle: crystal structure of putative haloacid dehalogenase-like hydrolase2 (yp_001086940.1) from clostridium difficile 630 at 2.10 a resolution
|
| 65 | d1wzca1 |
|
not modelled |
17.1 |
15 |
Fold:HAD-like
Superfamily:HAD-like
Family:Predicted hydrolases Cof |
| 66 | c3l8hC_ |
|
not modelled |
16.5 |
13 |
PDB header:hydrolase
Chain: C: PDB Molecule:putative haloacid dehalogenase-like hydrolase;
PDBTitle: crystal structure of d,d-heptose 1.7-bisphosphate phosphatase from b.2 bronchiseptica complexed with magnesium and phosphate
|
| 67 | d1wpga2 |
|
not modelled |
16.0 |
10 |
Fold:HAD-like
Superfamily:HAD-like
Family:Meta-cation ATPase, catalytic domain P |
| 68 | d1eo1a_ |
|
not modelled |
15.9 |
13 |
Fold:Ribonuclease H-like motif
Superfamily:Nitrogenase accessory factor-like
Family:MTH1175-like |
| 69 | d1ovma1 |
|
not modelled |
15.4 |
13 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 70 | d1ydha_ |
|
not modelled |
15.3 |
19 |
Fold:MCP/YpsA-like
Superfamily:MCP/YpsA-like
Family:MoCo carrier protein-like |
| 71 | d1k1ea_ |
|
not modelled |
15.3 |
13 |
Fold:HAD-like
Superfamily:HAD-like
Family:Probable phosphatase YrbI |
| 72 | d1wmaa1 |
|
not modelled |
14.7 |
29 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 73 | c3d54D_ |
|
not modelled |
13.7 |
25 |
PDB header:ligase
Chain: D: PDB Molecule:phosphoribosylformylglycinamidine synthase 1;
PDBTitle: stucture of purlqs from thermotoga maritima
|
| 74 | d2bdua1 |
|
not modelled |
13.5 |
11 |
Fold:HAD-like
Superfamily:HAD-like
Family:Pyrimidine 5'-nucleotidase (UMPH-1) |
| 75 | c2i55C_ |
|
not modelled |
12.7 |
14 |
PDB header:isomerase
Chain: C: PDB Molecule:phosphomannomutase;
PDBTitle: complex of glucose-1,6-bisphosphate with phosphomannomutase from2 leishmania mexicana
|
| 76 | d1gzda_ |
|
not modelled |
12.6 |
9 |
Fold:SIS domain
Superfamily:SIS domain
Family:Phosphoglucose isomerase, PGI |
| 77 | c3ewiB_ |
|
not modelled |
12.2 |
8 |
PDB header:transferase
Chain: B: PDB Molecule:n-acylneuraminate cytidylyltransferase;
PDBTitle: structural analysis of the c-terminal domain of murine cmp-2 sialic acid synthetase
|
| 78 | d2b3za1 |
|
not modelled |
11.6 |
13 |
Fold:Dihydrofolate reductase-like
Superfamily:Dihydrofolate reductase-like
Family:RibD C-terminal domain-like |
| 79 | d1a9xa3 |
|
not modelled |
11.5 |
15 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 80 | c3gh1A_ |
|
not modelled |
11.3 |
30 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:predicted nucleotide-binding protein;
PDBTitle: crystal structure of predicted nucleotide-binding protein from vibrio2 cholerae
|
| 81 | c3jtwB_ |
|
not modelled |
11.1 |
10 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:dihydrofolate reductase;
PDBTitle: crystal structure of putative dihydrofolate reductase (yp_805003.1)2 from pediococcus pentosaceus atcc 25745 at 1.90 a resolution
|
| 82 | d2obba1 |
|
not modelled |
11.1 |
9 |
Fold:HAD-like
Superfamily:HAD-like
Family:BT0820-like |
| 83 | d2azna1 |
|
not modelled |
10.7 |
10 |
Fold:Dihydrofolate reductase-like
Superfamily:Dihydrofolate reductase-like
Family:RibD C-terminal domain-like |
| 84 | d1n57a_ |
|
not modelled |
10.7 |
14 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:DJ-1/PfpI |
| 85 | c3pr3B_ |
|
not modelled |
10.3 |
18 |
PDB header:isomerase
Chain: B: PDB Molecule:glucose-6-phosphate isomerase;
PDBTitle: crystal structure of plasmodium falciparum glucose-6-phosphate2 isomerase (pf14_0341) in complex with fructose-6-phosphate
|
| 86 | c3rlhA_ |
|
not modelled |
10.0 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:sphingomyelin phosphodiesterase d lisictox-alphaia1a;
PDBTitle: crystal structure of a class ii phospholipase d from loxosceles2 intermedia venom
|
| 87 | d1rdua_ |
|
not modelled |
10.0 |
8 |
Fold:Ribonuclease H-like motif
Superfamily:Nitrogenase accessory factor-like
Family:MTH1175-like |
| 88 | d1iata_ |
|
not modelled |
9.9 |
9 |
Fold:SIS domain
Superfamily:SIS domain
Family:Phosphoglucose isomerase, PGI |
| 89 | d1l6ra_ |
|
not modelled |
9.7 |
16 |
Fold:HAD-like
Superfamily:HAD-like
Family:Predicted hydrolases Cof |
| 90 | c2qtdA_ |
|
not modelled |
9.4 |
6 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:uncharacterized protein mj0327;
PDBTitle: crystal structure of a putative dinitrogenase (mj0327) from2 methanocaldococcus jannaschii dsm at 1.70 a resolution
|
| 91 | c2qbuA_ |
|
not modelled |
9.4 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:precorrin-2 methyltransferase;
PDBTitle: crystal structure of methanothermobacter thermautotrophicus cbil
|
| 92 | c3rlgA_ |
|
not modelled |
9.3 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:sphingomyelin phosphodiesterase d lisictox-alphaia1a;
PDBTitle: crystal structure of loxosceles intermedia phospholipase d isoform 12 h12a mutant
|
| 93 | c3k3pA_ |
|
not modelled |
9.2 |
20 |
PDB header:ligase
Chain: A: PDB Molecule:d-alanine--d-alanine ligase;
PDBTitle: crystal structure of the apo form of d-alanine:d-alanine ligase (ddl)2 from streptococcus mutans
|
| 94 | d1r0ka3 |
|
not modelled |
9.0 |
10 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:Dihydrodipicolinate reductase-like |
| 95 | c3ujhB_ |
|
not modelled |
8.9 |
22 |
PDB header:isomerase
Chain: B: PDB Molecule:glucose-6-phosphate isomerase;
PDBTitle: crystal structure of substrate-bound glucose-6-phosphate isomerase2 from toxoplasma gondii
|
| 96 | c2o2cB_ |
|
not modelled |
8.8 |
18 |
PDB header:isomerase
Chain: B: PDB Molecule:glucose-6-phosphate isomerase, glycosomal;
PDBTitle: crystal structure of phosphoglucose isomerase from t. brucei2 containing glucose-6-phosphate in the active site
|
| 97 | c3nbuC_ |
|
not modelled |
8.6 |
16 |
PDB header:isomerase
Chain: C: PDB Molecule:glucose-6-phosphate isomerase;
PDBTitle: crystal structure of pgi glucosephosphate isomerase
|
| 98 | c2ekdD_ |
|
not modelled |
8.5 |
25 |
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:hypothetical protein ph0250;
PDBTitle: structural study of project id ph0250 from pyrococcus horikoshii ot3
|
| 99 | d1o8ca2 |
|
not modelled |
8.4 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Alcohol dehydrogenase-like, C-terminal domain |