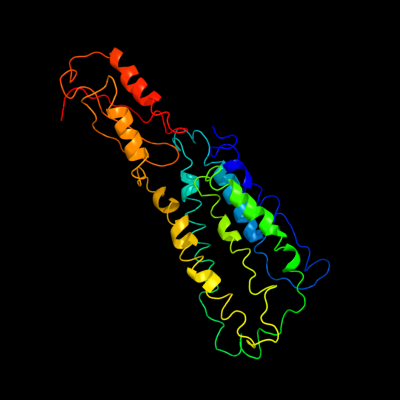
1 c2qfiB_
100.0
100
PDB header: transport proteinChain: B: PDB Molecule: ferrous-iron efflux pump fief;PDBTitle: structure of the zinc transporter yiip
2 d2qfia2
100.0
100
Fold: Cation efflux protein transmembrane domain-likeSuperfamily: Cation efflux protein transmembrane domain-likeFamily: Cation efflux protein transmembrane domain-like3 c2zztA_
99.6
25
PDB header: transport proteinChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of the cytosolic domain of the cation2 diffusion facilitator family protein
4 d3bypa1
99.6
34
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Cation efflux protein cytoplasmic domain-likeFamily: Cation efflux protein cytoplasmic domain-like5 d2qfia1
99.5
100
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Cation efflux protein cytoplasmic domain-likeFamily: Cation efflux protein cytoplasmic domain-like6 d1dlca3
58.5
18
Fold: Toxins' membrane translocation domainsSuperfamily: delta-Endotoxin (insectocide), N-terminal domainFamily: delta-Endotoxin (insectocide), N-terminal domain7 d1ghha_
57.6
15
Fold: DNA damage-inducible protein DinISuperfamily: DNA damage-inducible protein DinIFamily: DNA damage-inducible protein DinI8 c3qovD_
56.2
9
PDB header: ligaseChain: D: PDB Molecule: phenylacetate-coenzyme a ligase;PDBTitle: crystal structure of a hypothetical acyl-coa ligase (bt_0428) from2 bacteroides thetaiotaomicron vpi-5482 at 2.20 a resolution
9 c2y4oA_
53.7
16
PDB header: ligaseChain: A: PDB Molecule: phenylacetate-coenzyme a ligase;PDBTitle: crystal structure of paak2 in complex with phenylacetyl adenylate
10 c3cd0B_
53.3
14
PDB header: oxidoreductaseChain: B: PDB Molecule: 3-hydroxy-3-methylglutaryl-coenzyme a reductase;PDBTitle: thermodynamic and structure guided design of statin hmg-coa2 reductase inhibitors
11 d1ji6a3
45.8
15
Fold: Toxins' membrane translocation domainsSuperfamily: delta-Endotoxin (insectocide), N-terminal domainFamily: delta-Endotoxin (insectocide), N-terminal domain12 c1hwjB_
45.5
14
PDB header: oxidoreductaseChain: B: PDB Molecule: hmg-coa reductase;PDBTitle: complex of the catalytic portion of human hmg-coa reductase2 with cerivastatin
13 c1ji6A_
42.2
16
PDB header: toxinChain: A: PDB Molecule: pesticidial crystal protein cry3bb;PDBTitle: crystal structure of the insecticidal bacterial del2 endotoxin cry3bb1 bacillus thuringiensis
14 c2y27B_
41.8
12
PDB header: ligaseChain: B: PDB Molecule: phenylacetate-coenzyme a ligase;PDBTitle: crystal structure of paak1 in complex with atp from2 burkholderia cenocepacia
15 c3rfuC_
39.6
11
PDB header: hydrolase, membrane proteinChain: C: PDB Molecule: copper efflux atpase;PDBTitle: crystal structure of a copper-transporting pib-type atpase
16 d1f06a2
35.8
19
Fold: FwdE/GAPDH domain-likeSuperfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domainFamily: Dihydrodipicolinate reductase-like17 d1dqaa1
35.5
14
Fold: Ferredoxin-likeSuperfamily: NAD-binding domain of HMG-CoA reductaseFamily: NAD-binding domain of HMG-CoA reductase18 c3eb7B_
32.0
15
PDB header: toxinChain: B: PDB Molecule: insecticidal delta-endotoxin cry8ea1;PDBTitle: crystal structure of insecticidal delta-endotoxin cry8ea1 from2 bacillus thuringiensis at 2.2 angstroms resolution
19 c1dlcA_
31.5
18
PDB header: toxinChain: A: PDB Molecule: delta-endotoxin cryiiia;PDBTitle: crystal structure of insecticidal delta-endotoxin from2 bacillus thuringiensis at 2.5 angstroms resolution
20 d1wh9a_
31.3
12
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Prokaryotic type KH domain (KH-domain type II)Family: Prokaryotic type KH domain (KH-domain type II)21 d1ib8a2
not modelled
29.7
8
Fold: Alpha-lytic protease prodomain-likeSuperfamily: YhbC-like, N-terminal domainFamily: YhbC-like, N-terminal domain22 d1u8sa1
not modelled
29.1
17
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Glycine cleavage system transcriptional repressor23 c2jo1A_
not modelled
27.0
9
PDB header: hydrolase regulatorChain: A: PDB Molecule: phospholemman;PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
24 c3b64A_
not modelled
24.4
13
PDB header: cytokineChain: A: PDB Molecule: macrophage migration inhibitory factor-likePDBTitle: macrophage migration inhibitory factor (mif) from2 /leishmania major
25 c1u8sB_
not modelled
21.7
15
PDB header: transcriptionChain: B: PDB Molecule: glycine cleavage system transcriptionalPDBTitle: crystal structure of putative glycine cleavage system2 transcriptional repressor
26 d1uwda_
not modelled
19.3
17
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Fe-S cluster assembly (FSCA) domain-likeFamily: PaaD-like27 c2jp3A_
not modelled
18.7
0
PDB header: transcriptionChain: A: PDB Molecule: fxyd domain-containing ion transport regulator 4;PDBTitle: solution structure of the human fxyd4 (chif) protein in sds2 micelles
28 c3abfB_
not modelled
17.4
9
PDB header: isomeraseChain: B: PDB Molecule: 4-oxalocrotonate tautomerase;PDBTitle: crystal structure of a 4-oxalocrotonate tautomerase homologue2 (tthb242)
29 d1fftb2
not modelled
16.8
15
Fold: Transmembrane helix hairpinSuperfamily: Cytochrome c oxidase subunit II-like, transmembrane regionFamily: Cytochrome c oxidase subunit II-like, transmembrane region30 d1zpva1
not modelled
16.7
6
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: SP0238-like31 c3laxA_
not modelled
16.3
6
PDB header: ligaseChain: A: PDB Molecule: phenylacetate-coenzyme a ligase;PDBTitle: the crystal structure of a domain of phenylacetate-coenzyme2 a ligase from bacteroides vulgatus atcc 8482
32 c3eynB_
not modelled
16.1
8
PDB header: ligaseChain: B: PDB Molecule: acyl-coenzyme a synthetase acsm2a;PDBTitle: crystal structure of human acyl-coa synthetase medium-chain2 family member 2a (l64p mutation) in a complex with coa
33 d1u8sa2
not modelled
15.5
14
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Glycine cleavage system transcriptional repressor34 c3etcB_
not modelled
14.8
7
PDB header: ligaseChain: B: PDB Molecule: amp-binding protein;PDBTitle: 2.1 a structure of acyl-adenylate synthetase from methanosarcina2 acetivorans containing a link between lys256 and cys298
35 c2zxeG_
not modelled
12.8
6
PDB header: hydrolase/transport proteinChain: G: PDB Molecule: phospholemman-like protein;PDBTitle: crystal structure of the sodium - potassium pump in the e2.2k+.pi2 state
36 d1x38a2
not modelled
11.7
25
Fold: Flavodoxin-likeSuperfamily: Beta-D-glucan exohydrolase, C-terminal domainFamily: Beta-D-glucan exohydrolase, C-terminal domain37 c2ns6A_
not modelled
11.7
17
PDB header: hydrolaseChain: A: PDB Molecule: mobilization protein a;PDBTitle: crystal structure of the minimal relaxase domain of moba2 from plasmid r1162
38 c3kxwA_
not modelled
11.6
9
PDB header: ligaseChain: A: PDB Molecule: saframycin mx1 synthetase b;PDBTitle: the crystal structure of fatty acid amp ligase from legionella2 pneumophila
39 c3hvwA_
not modelled
11.1
13
PDB header: lyaseChain: A: PDB Molecule: diguanylate-cyclase (dgc);PDBTitle: crystal structure of the ggdef domain of the pa2567 protein2 from pseudomonas aeruginosa, northeast structural genomics3 consortium target par365c
40 c2iqcA_
not modelled
9.8
14
PDB header: protein bindingChain: A: PDB Molecule: fanconi anemia group f protein;PDBTitle: crystal structure of human fancf protein that functions in2 the assembly of a dna damage signaling complex
41 d1q8ba_
not modelled
9.8
10
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: Hypothetical protein YjcS42 c3gacD_
not modelled
9.7
23
PDB header: cytokineChain: D: PDB Molecule: macrophage migration inhibitory factor-likePDBTitle: structure of mif with hpp
43 d2cu6a1
not modelled
9.5
16
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Fe-S cluster assembly (FSCA) domain-likeFamily: PaaD-like44 d1rq2a2
not modelled
9.4
18
Fold: Bacillus chorismate mutase-likeSuperfamily: Tubulin C-terminal domain-likeFamily: Tubulin, C-terminal domain45 d1mlia_
not modelled
9.4
5
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: Muconalactone isomerase, MLI46 d2aala1
not modelled
9.2
13
Fold: Tautomerase/MIFSuperfamily: Tautomerase/MIFFamily: MSAD-like47 c2x4kB_
not modelled
8.9
7
PDB header: isomeraseChain: B: PDB Molecule: 4-oxalocrotonate tautomerase;PDBTitle: crystal structure of sar1376, a putative 4-oxalocrotonate2 tautomerase from the methicillin-resistant staphylococcus3 aureus (mrsa)
48 d1wzua1
not modelled
8.8
19
Fold: NadA-likeSuperfamily: NadA-likeFamily: NadA-like49 c3mb2G_
not modelled
8.3
16
PDB header: isomeraseChain: G: PDB Molecule: 4-oxalocrotonate tautomerase family enzyme - alpha subunit;PDBTitle: kinetic and structural characterization of a heterohexamer 4-2 oxalocrotonate tautomerase from chloroflexus aurantiacus j-10-fl:3 implications for functional and structural diversity in the4 tautomerase superfamily
50 c1ex1A_
not modelled
7.9
25
PDB header: hydrolaseChain: A: PDB Molecule: protein (beta-d-glucan exohydrolase isoenzyme exo1);PDBTitle: beta-d-glucan exohydrolase from barley
51 d1lcia_
not modelled
7.7
11
Fold: Acetyl-CoA synthetase-likeSuperfamily: Acetyl-CoA synthetase-likeFamily: Acetyl-CoA synthetase-like52 d1rwua_
not modelled
7.7
8
Fold: Ferredoxin-likeSuperfamily: YbeD/HP0495-likeFamily: YbeD-like53 c1rwuA_
not modelled
7.7
8
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical upf0250 protein ybed;PDBTitle: solution structure of conserved protein ybed from e. coli
54 c3e7wA_
not modelled
7.4
14
PDB header: ligaseChain: A: PDB Molecule: d-alanine--poly(phosphoribitol) ligase subunit 1;PDBTitle: crystal structure of dlta: implications for the reaction2 mechanism of non-ribosomal peptide synthetase (nrps)3 adenylation domains
55 c1v55B_
not modelled
7.3
9
PDB header: oxidoreductaseChain: B: PDB Molecule: cytochrome c oxidase polypeptide ii;PDBTitle: bovine heart cytochrome c oxidase at the fully reduced state
56 c3mlcC_
not modelled
7.2
19
PDB header: isomeraseChain: C: PDB Molecule: fg41 malonate semialdehyde decarboxylase;PDBTitle: crystal structure of fg41msad inactivated by 3-chloropropiolate
57 c3m20A_
not modelled
7.0
10
PDB header: isomeraseChain: A: PDB Molecule: 4-oxalocrotonate tautomerase, putative;PDBTitle: crystal structure of dmpi from archaeoglobus fulgidus determined to2 2.37 angstroms resolution
58 d1uf0a_
not modelled
6.8
8
Fold: beta-Grasp (ubiquitin-like)Superfamily: Doublecortin (DC)Family: Doublecortin (DC)59 d1xhja_
not modelled
6.8
11
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Fe-S cluster assembly (FSCA) domain-likeFamily: NifU C-terminal domain-like60 c2qs0A_
not modelled
6.7
14
PDB header: biosynthetic proteinChain: A: PDB Molecule: quinolinate synthetase a;PDBTitle: quinolinate synthase from pyrococcus furiosus
61 c3ry0A_
not modelled
6.6
10
PDB header: isomeraseChain: A: PDB Molecule: putative tautomerase;PDBTitle: crystal structure of tomn, a 4-oxalocrotonate tautomerase homologue in2 tomaymycin biosynthetic pathway
62 c2zkqc_
not modelled
6.5
13
PDB header: ribosomal protein/rnaChain: C: PDB Molecule: rna expansion segment es4;PDBTitle: structure of a mammalian ribosomal 40s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
63 c3dptB_
not modelled
6.4
17
PDB header: signaling proteinChain: B: PDB Molecule: rab family protein;PDBTitle: cor domain of rab family protein (roco)
64 c1r7iB_
not modelled
6.4
14
PDB header: oxidoreductaseChain: B: PDB Molecule: 3-hydroxy-3-methylglutaryl-coenzyme a reductase;PDBTitle: hmg-coa reductase from p. mevalonii, native structure at 2.2 angstroms2 resolution.
65 c2kz0A_
not modelled
6.4
26
PDB header: transcriptionChain: A: PDB Molecule: bola family protein;PDBTitle: solution structure of a bola protein (ech_0303) from ehrlichia2 chaffeensis. seattle structural genomics center for infectious3 disease target ehcha.10365.a
66 c3ibwA_
not modelled
6.3
3
PDB header: transferaseChain: A: PDB Molecule: gtp pyrophosphokinase;PDBTitle: crystal structure of the act domain from gtp2 pyrophosphokinase of chlorobium tepidum. northeast3 structural genomics consortium target ctr148a
67 c2zw2B_
not modelled
6.3
15
PDB header: ligaseChain: B: PDB Molecule: putative uncharacterized protein sts178;PDBTitle: crystal structure of formylglycinamide ribonucleotide amidotransferase2 iii from sulfolobus tokodaii (stpurs)
68 c3nyrA_
not modelled
6.3
18
PDB header: ligaseChain: A: PDB Molecule: malonyl-coa ligase;PDBTitle: malonyl-coa ligase ternary product complex with malonyl-coa and amp2 bound
69 c2dbbA_
not modelled
6.2
11
PDB header: transcriptional regulatorChain: A: PDB Molecule: putative hth-type transcriptional regulator ph0061;PDBTitle: crystal structure of ph0061
70 d1uxya2
not modelled
6.1
20
Fold: Uridine diphospho-N-Acetylenolpyruvylglucosamine reductase, MurB, C-terminal domainSuperfamily: Uridine diphospho-N-Acetylenolpyruvylglucosamine reductase, MurB, C-terminal domainFamily: Uridine diphospho-N-Acetylenolpyruvylglucosamine reductase, MurB, C-terminal domain71 d2cfxa2
not modelled
6.1
3
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: Lrp/AsnC-like transcriptional regulator C-terminal domain72 c2qieA_
not modelled
6.1
10
PDB header: transferaseChain: A: PDB Molecule: molybdopterin-converting factor subunit 2;PDBTitle: staphylococcus aureus molybdopterin synthase in complex2 with precursor z
73 d1iwga3
not modelled
6.0
8
Fold: Ferredoxin-likeSuperfamily: Multidrug efflux transporter AcrB pore domain; PN1, PN2, PC1 and PC2 subdomainsFamily: Multidrug efflux transporter AcrB pore domain; PN1, PN2, PC1 and PC2 subdomains74 c3lnoA_
not modelled
6.0
11
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of domain of unknown function duf59 from2 bacillus anthracis
75 d1gyxa_
not modelled
6.0
18
Fold: Tautomerase/MIFSuperfamily: Tautomerase/MIFFamily: 4-oxalocrotonate tautomerase-like76 c3ac0B_
not modelled
6.0
25
PDB header: hydrolaseChain: B: PDB Molecule: beta-glucosidase i;PDBTitle: crystal structure of beta-glucosidase from kluyveromyces marxianus in2 complex with glucose
77 d2vapa2
not modelled
5.9
28
Fold: Bacillus chorismate mutase-likeSuperfamily: Tubulin C-terminal domain-likeFamily: Tubulin, C-terminal domain78 d2ia7a1
not modelled
5.8
7
Fold: gpW/gp25-likeSuperfamily: gpW/gp25-likeFamily: gpW/gp25-like79 c2op8A_
not modelled
5.8
9
PDB header: isomeraseChain: A: PDB Molecule: probable tautomerase ywhb;PDBTitle: crystal structure of ywhb- homologue of 4-oxalocrotonate tautomerase
80 d1ofua2
not modelled
5.7
21
Fold: Bacillus chorismate mutase-likeSuperfamily: Tubulin C-terminal domain-likeFamily: Tubulin, C-terminal domain81 d2b3ja1
not modelled
5.7
12
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Deoxycytidylate deaminase-like82 c2xq2A_
not modelled
5.7
17
PDB header: transport proteinChain: A: PDB Molecule: sodium/glucose cotransporter;PDBTitle: structure of the k294a mutant of vsglt
83 d2i1sa1
not modelled
5.6
15
Fold: MM3350-likeSuperfamily: MM3350-likeFamily: MM3350-like84 c2x41A_
not modelled
5.6
25
PDB header: hydrolaseChain: A: PDB Molecule: beta-glucosidase;PDBTitle: structure of beta-glucosidase 3b from thermotoga neapolitana2 in complex with glucose
85 c3brtC_
not modelled
5.5
29
PDB header: transferase/transcriptionChain: C: PDB Molecule: inhibitor of nuclear factor kappa-b kinasePDBTitle: nemo/ikk association domain structure
86 c2e1cA_
not modelled
5.4
10
PDB header: transcription/dnaChain: A: PDB Molecule: putative hth-type transcriptional regulator ph1519;PDBTitle: structure of putative hth-type transcriptional regulator ph1519/dna2 complex
87 d1otfa_
not modelled
5.4
18
Fold: Tautomerase/MIFSuperfamily: Tautomerase/MIFFamily: 4-oxalocrotonate tautomerase-like88 d1hp3a_
not modelled
5.4
31
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: omega toxin-likeFamily: Spider toxins89 c3dhvA_
not modelled
5.4
11
PDB header: ligaseChain: A: PDB Molecule: d-alanine-poly(phosphoribitol) ligase;PDBTitle: crystal structure of dlta protein in complex with d-alanine2 adenylate
90 c3gdzA_
not modelled
5.3
14
PDB header: ligaseChain: A: PDB Molecule: arginyl-trna synthetase;PDBTitle: crystal structure of arginyl-trna synthetase from klebsiella2 pneumoniae subsp. pneumoniae
91 c1s1hC_
not modelled
5.3
13
PDB header: ribosomeChain: C: PDB Molecule: 40s ribosomal protein s3;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1h,4 contains 40s subunit. the 60s ribosomal subunit is in file5 1s1i.
92 c1ypxA_
not modelled
5.3
15
PDB header: transferaseChain: A: PDB Molecule: putative vitamin-b12 independent methionine synthase familyPDBTitle: crystal structure of the putative vitamin-b12 independent methionine2 synthase from listeria monocytogenes, northeast structural genomics3 target lmr13
93 c2vv5D_
not modelled
5.3
14
PDB header: membrane proteinChain: D: PDB Molecule: small-conductance mechanosensitive channel;PDBTitle: the open structure of mscs
94 d2atza1
not modelled
5.3
10
Fold: Prim-pol domainSuperfamily: Prim-pol domainFamily: HP0184-like95 c1ib8A_
not modelled
5.3
12
PDB header: nucleic acid binding proteinChain: A: PDB Molecule: conserved protein sp14.3;PDBTitle: solution structure and function of a conserved protein2 sp14.3 encoded by an essential streptococcus pneumoniae3 gene
96 c2khuA_
not modelled
5.2
7
PDB header: transferase/protein bindingChain: A: PDB Molecule: immunoglobulin g-binding protein g, dnaPDBTitle: solution structure of the ubiquitin-binding motif of human2 polymerase iota
97 c3fwtA_
not modelled
5.0
10
PDB header: cytokineChain: A: PDB Molecule: macrophage migration inhibitory factor-likePDBTitle: crystal structure of leishmania major mif2
98 c3lk6A_
not modelled
5.0
27
PDB header: hydrolaseChain: A: PDB Molecule: lipoprotein ybbd;PDBTitle: beta-n-hexosaminidase n318d mutant (ybbd_n318d) from bacillus subtilis
99 d1g9pa_
not modelled
5.0
31
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: omega toxin-likeFamily: Spider toxins