| 1 | c1kjjA_ |
|
 |
100.0 |
100 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase 2;
PDBTitle: crystal structure of glycniamide ribonucleotide2 transformylase in complex with mg-atp-gamma-s
|
| 2 | c2dwcB_ |
|
 |
100.0 |
49 |
PDB header:transferase
Chain: B: PDB Molecule:433aa long hypothetical phosphoribosylglycinamide formyl
PDBTitle: crystal structure of probable phosphoribosylglycinamide formyl2 transferase from pyrococcus horikoshii ot3 complexed with adp
|
| 3 | c3uvzB_ |
|
 |
100.0 |
24 |
PDB header:lyase
Chain: B: PDB Molecule:phosphoribosylaminoimidazole carboxylase, atpase subunit;
PDBTitle: crystal structure of phosphoribosylaminoimidazole carboxylase, atpase2 subunit from burkholderia ambifaria
|
| 4 | c3q2oB_ |
|
 |
100.0 |
26 |
PDB header:lyase
Chain: B: PDB Molecule:phosphoribosylaminoimidazole carboxylase, atpase subunit;
PDBTitle: crystal structure of purk: n5-carboxyaminoimidazole ribonucleotide2 synthetase
|
| 5 | c3orqA_ |
|
 |
100.0 |
21 |
PDB header:ligase,biosynthetic protein
Chain: A: PDB Molecule:n5-carboxyaminoimidazole ribonucleotide synthetase;
PDBTitle: crystal structure of n5-carboxyaminoimidazole synthetase from2 staphylococcus aureus complexed with adp
|
| 6 | c3bg5C_ |
|
 |
100.0 |
19 |
PDB header:ligase
Chain: C: PDB Molecule:pyruvate carboxylase;
PDBTitle: crystal structure of staphylococcus aureus pyruvate2 carboxylase
|
| 7 | c3k5iB_ |
|
 |
100.0 |
23 |
PDB header:lyase
Chain: B: PDB Molecule:phosphoribosyl-aminoimidazole carboxylase;
PDBTitle: crystal structure of n5-carboxyaminoimidazole synthase from2 aspergillus clavatus in complex with adp and 5-3 aminoimadazole ribonucleotide
|
| 8 | c3g8cB_ |
|
 |
100.0 |
19 |
PDB header:ligase
Chain: B: PDB Molecule:biotin carboxylase;
PDBTitle: crystal stucture of biotin carboxylase in complex with2 biotin, bicarbonate, adp and mg ion
|
| 9 | c3etjB_ |
|
 |
100.0 |
22 |
PDB header:lyase
Chain: B: PDB Molecule:phosphoribosylaminoimidazole carboxylase atpase
PDBTitle: crystal structure e. coli purk in complex with mg, adp, and2 pi
|
| 10 | c3ouzA_ |
|
 |
100.0 |
16 |
PDB header:ligase
Chain: A: PDB Molecule:biotin carboxylase;
PDBTitle: crystal structure of biotin carboxylase-adp complex from campylobacter2 jejuni
|
| 11 | c1ulzA_ |
|
 |
100.0 |
16 |
PDB header:ligase
Chain: A: PDB Molecule:pyruvate carboxylase n-terminal domain;
PDBTitle: crystal structure of the biotin carboxylase subunit of pyruvate2 carboxylase
|
| 12 | c2dzdB_ |
|
 |
100.0 |
19 |
PDB header:ligase
Chain: B: PDB Molecule:pyruvate carboxylase;
PDBTitle: crystal structure of the biotin carboxylase domain of2 pyruvate carboxylase
|
| 13 | c2vpqA_ |
|
 |
100.0 |
16 |
PDB header:ligase
Chain: A: PDB Molecule:acetyl-coa carboxylase;
PDBTitle: crystal structure of biotin carboxylase from s. aureus2 complexed with amppnp
|
| 14 | c2yyaB_ |
|
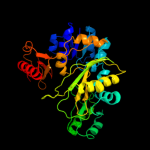 |
100.0 |
17 |
PDB header:ligase
Chain: B: PDB Molecule:phosphoribosylamine--glycine ligase;
PDBTitle: crystal structure of gar synthetase from aquifex aeolicus
|
| 15 | c1m6vE_ |
|
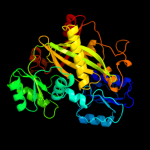 |
100.0 |
16 |
PDB header:ligase
Chain: E: PDB Molecule:carbamoyl phosphate synthetase large chain;
PDBTitle: crystal structure of the g359f (small subunit) point mutant of2 carbamoyl phosphate synthetase
|
| 16 | c2hjwA_ |
|
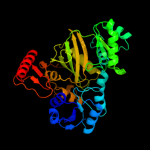 |
100.0 |
18 |
PDB header:ligase
Chain: A: PDB Molecule:acetyl-coa carboxylase 2;
PDBTitle: crystal structure of the bc domain of acc2
|
| 17 | c2gpwC_ |
|
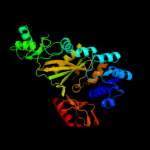 |
100.0 |
19 |
PDB header:ligase
Chain: C: PDB Molecule:biotin carboxylase;
PDBTitle: crystal structure of the biotin carboxylase subunit, f363a2 mutant, of acetyl-coa carboxylase from escherichia coli.
|
| 18 | c1w96B_ |
|
 |
100.0 |
14 |
PDB header:ligase
Chain: B: PDB Molecule:acetyl-coenzyme a carboxylase;
PDBTitle: crystal structure of biotin carboxylase domain of acetyl-2 coenzyme a carboxylase from saccharomyces cerevisiae in3 complex with soraphen a
|
| 19 | c2xd4A_ |
|
 |
100.0 |
17 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylamine--glycine ligase;
PDBTitle: nucleotide-bound structures of bacillus subtilis glycinamide2 ribonucleotide synthetase
|
| 20 | c3lp8A_ |
|
 |
100.0 |
18 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylamine-glycine ligase;
PDBTitle: crystal structure of phosphoribosylamine-glycine ligase from2 ehrlichia chaffeensis
|
| 21 | c3gidB_ |
|
not modelled |
100.0 |
17 |
PDB header:ligase
Chain: B: PDB Molecule:acetyl-coa carboxylase 2;
PDBTitle: the biotin carboxylase (bc) domain of human acetyl-coa2 carboxylase 2 (acc2) in complex with soraphen a
|
| 22 | c3n6rK_ |
|
not modelled |
100.0 |
17 |
PDB header:ligase
Chain: K: PDB Molecule:propionyl-coa carboxylase, alpha subunit;
PDBTitle: crystal structure of the holoenzyme of propionyl-coa carboxylase (pcc)
|
| 23 | c3u9sE_ |
|
not modelled |
100.0 |
20 |
PDB header:ligase
Chain: E: PDB Molecule:methylcrotonyl-coa carboxylase, alpha-subunit;
PDBTitle: crystal structure of p. aeruginosa 3-methylcrotonyl-coa carboxylase2 (mcc) 750 kd holoenzyme, coa complex
|
| 24 | c2qk4A_ |
|
not modelled |
100.0 |
17 |
PDB header:ligase
Chain: A: PDB Molecule:trifunctional purine biosynthetic protein adenosine-3;
PDBTitle: human glycinamide ribonucleotide synthetase
|
| 25 | c2ys6A_ |
|
not modelled |
100.0 |
18 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylglycinamide synthetase;
PDBTitle: crystal structure of gar synthetase from geobacillus kaustophilus
|
| 26 | c2ip4A_ |
|
not modelled |
100.0 |
20 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylamine--glycine ligase;
PDBTitle: crystal structure of glycinamide ribonucleotide synthetase from2 thermus thermophilus hb8
|
| 27 | c1vkzA_ |
|
not modelled |
100.0 |
16 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylamine--glycine ligase;
PDBTitle: crystal structure of phosphoribosylamine--glycine ligase (tm1250) from2 thermotoga maritima at 2.30 a resolution
|
| 28 | c2z04A_ |
|
not modelled |
100.0 |
28 |
PDB header:lyase
Chain: A: PDB Molecule:phosphoribosylaminoimidazole carboxylase atpase
PDBTitle: crystal structure of phosphoribosylaminoimidazole2 carboxylase atpase subunit from aquifex aeolicus
|
| 29 | c1gsoA_ |
|
not modelled |
100.0 |
17 |
PDB header:ligase
Chain: A: PDB Molecule:protein (glycinamide ribonucleotide synthetase);
PDBTitle: glycinamide ribonucleotide synthetase (gar-syn) from e.2 coli.
|
| 30 | d1a9xa5 |
|
not modelled |
100.0 |
12 |
Fold:ATP-grasp
Superfamily:Glutathione synthetase ATP-binding domain-like
Family:BC ATP-binding domain-like |
| 31 | c2qf7A_ |
|
not modelled |
100.0 |
21 |
PDB header:ligase
Chain: A: PDB Molecule:pyruvate carboxylase protein;
PDBTitle: crystal structure of a complete multifunctional pyruvate carboxylase2 from rhizobium etli
|
| 32 | c2i80B_ |
|
not modelled |
100.0 |
17 |
PDB header:ligase
Chain: B: PDB Molecule:d-alanine-d-alanine ligase;
PDBTitle: allosteric inhibition of staphylococcus aureus d-alanine:d-alanine2 ligase revealed by crystallographic studies
|
| 33 | c3lwbA_ |
|
not modelled |
100.0 |
23 |
PDB header:ligase
Chain: A: PDB Molecule:d-alanine--d-alanine ligase;
PDBTitle: crystal structure of apo d-alanine:d-alanine ligase (ddl) from2 mycobacterium tuberculosis
|
| 34 | c2pn1A_ |
|
not modelled |
100.0 |
16 |
PDB header:ligase
Chain: A: PDB Molecule:carbamoylphosphate synthase large subunit;
PDBTitle: crystal structure of carbamoylphosphate synthase large subunit (split2 gene in mj) (zp_00538348.1) from exiguobacterium sp. 255-15 at 2.00 a3 resolution
|
| 35 | c3i12A_ |
|
not modelled |
100.0 |
20 |
PDB header:ligase
Chain: A: PDB Molecule:d-alanine-d-alanine ligase a;
PDBTitle: the crystal structure of the d-alanyl-alanine synthetase a from2 salmonella enterica subsp. enterica serovar typhimurium str. lt2
|
| 36 | c1ehiB_ |
|
not modelled |
100.0 |
16 |
PDB header:ligase
Chain: B: PDB Molecule:d-alanine:d-lactate ligase;
PDBTitle: d-alanine:d-lactate ligase (lmddl2) of vancomycin-resistant2 leuconostoc mesenteroides
|
| 37 | d1kjqa3 |
|
not modelled |
100.0 |
87 |
Fold:ATP-grasp
Superfamily:Glutathione synthetase ATP-binding domain-like
Family:BC ATP-binding domain-like |
| 38 | c2dlnA_ |
|
not modelled |
100.0 |
19 |
PDB header:ligase(peptidoglycan synthesis)
Chain: A: PDB Molecule:d-alanine--d-alanine ligase;
PDBTitle: vancomycin resistance: structure of d-alanine:d-alanine2 ligase at 2.3 angstroms resolution
|
| 39 | c2r85B_ |
|
not modelled |
100.0 |
17 |
PDB header:unknown function
Chain: B: PDB Molecule:purp protein pf1517;
PDBTitle: crystal structure of purp from pyrococcus furiosus complexed with amp
|
| 40 | c1e4eB_ |
|
not modelled |
100.0 |
19 |
PDB header:ligase
Chain: B: PDB Molecule:vancomycin/teicoplanin a-type resistance protein vana;
PDBTitle: d-alanyl-d-lacate ligase
|
| 41 | c3r23B_ |
|
not modelled |
100.0 |
13 |
PDB header:ligase
Chain: B: PDB Molecule:d-alanine--d-alanine ligase;
PDBTitle: crystal structure of d-alanine--d-alanine ligase from bacillus2 anthracis
|
| 42 | d1w96a3 |
|
not modelled |
100.0 |
14 |
Fold:ATP-grasp
Superfamily:Glutathione synthetase ATP-binding domain-like
Family:BC ATP-binding domain-like |
| 43 | c2zdqA_ |
|
not modelled |
100.0 |
19 |
PDB header:ligase
Chain: A: PDB Molecule:d-alanine--d-alanine ligase;
PDBTitle: crystal structure of d-alanine:d-alanine ligase with atp2 and d-alanine:d-alanine from thermus thermophius hb8
|
| 44 | d1a9xa6 |
|
not modelled |
100.0 |
19 |
Fold:ATP-grasp
Superfamily:Glutathione synthetase ATP-binding domain-like
Family:BC ATP-binding domain-like |
| 45 | c3tqtB_ |
|
not modelled |
100.0 |
14 |
PDB header:ligase
Chain: B: PDB Molecule:d-alanine--d-alanine ligase;
PDBTitle: structure of the d-alanine-d-alanine ligase from coxiella burnetii
|
| 46 | c3e5nA_ |
|
not modelled |
100.0 |
22 |
PDB header:ligase
Chain: A: PDB Molecule:d-alanine-d-alanine ligase a;
PDBTitle: crystal strucutre of d-alanine-d-alanine ligase from2 xanthomonas oryzae pv. oryzae kacc10331
|
| 47 | c3se7A_ |
|
not modelled |
100.0 |
22 |
PDB header:ligase
Chain: A: PDB Molecule:vana;
PDBTitle: ancient vana
|
| 48 | c2pvpB_ |
|
not modelled |
100.0 |
18 |
PDB header:ligase
Chain: B: PDB Molecule:d-alanine-d-alanine ligase;
PDBTitle: crystal structure of d-alanine-d-alanine ligase from helicobacter2 pylori
|
| 49 | d3etja3 |
|
not modelled |
100.0 |
26 |
Fold:ATP-grasp
Superfamily:Glutathione synthetase ATP-binding domain-like
Family:BC ATP-binding domain-like |
| 50 | d2j9ga3 |
|
not modelled |
100.0 |
17 |
Fold:ATP-grasp
Superfamily:Glutathione synthetase ATP-binding domain-like
Family:BC ATP-binding domain-like |
| 51 | c3k3pA_ |
|
not modelled |
100.0 |
16 |
PDB header:ligase
Chain: A: PDB Molecule:d-alanine--d-alanine ligase;
PDBTitle: crystal structure of the apo form of d-alanine:d-alanine ligase (ddl)2 from streptococcus mutans
|
| 52 | d1vkza3 |
|
not modelled |
100.0 |
16 |
Fold:ATP-grasp
Superfamily:Glutathione synthetase ATP-binding domain-like
Family:BC ATP-binding domain-like |
| 53 | c3df7A_ |
|
not modelled |
100.0 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative atp-grasp superfamily protein;
PDBTitle: crystal structure of a putative atp-grasp superfamily2 protein from archaeoglobus fulgidus
|
| 54 | d1ulza3 |
|
not modelled |
100.0 |
16 |
Fold:ATP-grasp
Superfamily:Glutathione synthetase ATP-binding domain-like
Family:BC ATP-binding domain-like |
| 55 | c1uc8B_ |
|
not modelled |
100.0 |
17 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:lysine biosynthesis enzyme;
PDBTitle: crystal structure of a lysine biosynthesis enzyme, lysx,2 from thermus thermophilus hb8
|
| 56 | d2r7ka2 |
|
not modelled |
99.9 |
19 |
Fold:ATP-grasp
Superfamily:Glutathione synthetase ATP-binding domain-like
Family:PurP ATP-binding domain-like |
| 57 | d2r85a2 |
|
not modelled |
99.9 |
22 |
Fold:ATP-grasp
Superfamily:Glutathione synthetase ATP-binding domain-like
Family:PurP ATP-binding domain-like |
| 58 | d1ehia2 |
|
not modelled |
99.9 |
17 |
Fold:ATP-grasp
Superfamily:Glutathione synthetase ATP-binding domain-like
Family:ATP-binding domain of peptide synthetases |
| 59 | d1e4ea2 |
|
not modelled |
99.9 |
22 |
Fold:ATP-grasp
Superfamily:Glutathione synthetase ATP-binding domain-like
Family:ATP-binding domain of peptide synthetases |
| 60 | d1iowa2 |
|
not modelled |
99.9 |
19 |
Fold:ATP-grasp
Superfamily:Glutathione synthetase ATP-binding domain-like
Family:ATP-binding domain of peptide synthetases |
| 61 | d1gsoa3 |
|
not modelled |
99.9 |
14 |
Fold:ATP-grasp
Superfamily:Glutathione synthetase ATP-binding domain-like
Family:BC ATP-binding domain-like |
| 62 | d1uc8a2 |
|
not modelled |
99.9 |
16 |
Fold:ATP-grasp
Superfamily:Glutathione synthetase ATP-binding domain-like
Family:Lysine biosynthesis enzyme LysX ATP-binding domain |
| 63 | d1kjqa2 |
|
not modelled |
99.9 |
100 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 64 | c3ln6A_ |
|
not modelled |
99.9 |
16 |
PDB header:ligase
Chain: A: PDB Molecule:glutathione biosynthesis bifunctional protein gshab;
PDBTitle: crystal structure of a bifunctional glutathione synthetase from2 streptococcus agalactiae
|
| 65 | c2p0aA_ |
|
not modelled |
99.9 |
14 |
PDB header:neuropeptide
Chain: A: PDB Molecule:synapsin-3;
PDBTitle: the crystal structure of human synapsin iii (syn3) in complex with2 amppnp
|
| 66 | c1pk8D_ |
|
not modelled |
99.9 |
14 |
PDB header:membrane protein
Chain: D: PDB Molecule:rat synapsin i;
PDBTitle: crystal structure of rat synapsin i c domain complexed to2 ca.atp
|
| 67 | c1i7nA_ |
|
not modelled |
99.9 |
12 |
PDB header:neuropeptide
Chain: A: PDB Molecule:synapsin ii;
PDBTitle: crystal structure analysis of the c domain of synapsin ii2 from rat brain
|
| 68 | c1z2pX_ |
|
not modelled |
99.8 |
17 |
PDB header:transferase
Chain: X: PDB Molecule:inositol 1,3,4-trisphosphate 5/6-kinase;
PDBTitle: inositol 1,3,4-trisphosphate 5/6-kinase in complex with mg2+/amp-2 pcp/ins(1,3,4)p3
|
| 69 | c1gshA_ |
|
not modelled |
99.8 |
16 |
PDB header:glutathione biosynthesis ligase
Chain: A: PDB Molecule:glutathione biosynthetic ligase;
PDBTitle: structure of escherichia coli glutathione synthetase at ph 7.5
|
| 70 | d1pk8a2 |
|
not modelled |
99.8 |
12 |
Fold:ATP-grasp
Superfamily:Glutathione synthetase ATP-binding domain-like
Family:Synapsin C-terminal domain |
| 71 | c3ln7A_ |
|
not modelled |
99.8 |
18 |
PDB header:ligase
Chain: A: PDB Molecule:glutathione biosynthesis bifunctional protein gshab;
PDBTitle: crystal structure of a bifunctional glutathione synthetase from2 pasteurella multocida
|
| 72 | d1i7na2 |
|
not modelled |
99.8 |
12 |
Fold:ATP-grasp
Superfamily:Glutathione synthetase ATP-binding domain-like
Family:Synapsin C-terminal domain |
| 73 | c2qb5B_ |
|
not modelled |
99.8 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:inositol-tetrakisphosphate 1-kinase;
PDBTitle: crystal structure of human inositol 1,3,4-trisphosphate 5/6-kinase2 (itpk1) in complex with adp and mn2+
|
| 74 | d1gsaa2 |
|
not modelled |
99.6 |
9 |
Fold:ATP-grasp
Superfamily:Glutathione synthetase ATP-binding domain-like
Family:ATP-binding domain of peptide synthetases |
| 75 | c3t9aA_ |
|
not modelled |
99.5 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:inositol pyrophosphate kinase;
PDBTitle: crystal structure of the catalytic domain of human diphosphoinositol2 pentakisphosphate kinase 2 (ppip5k2) in complex with amppnp at ph 7.0
|
| 76 | c2r7mA_ |
|
not modelled |
99.5 |
17 |
PDB header:ligase
Chain: A: PDB Molecule:5-formaminoimidazole-4-carboxamide-1-(beta)-d-
PDBTitle: crystal structure of faicar synthetase (purp) from m.2 jannaschii complexed with amp
|
| 77 | c2cqyA_ |
|
not modelled |
99.5 |
17 |
PDB header:ligase
Chain: A: PDB Molecule:propionyl-coa carboxylase alpha chain,
PDBTitle: solution structure of b domain from human propionyl-coa2 carboxylase alpha subunit
|
| 78 | d1a9xa4 |
|
not modelled |
99.5 |
18 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 79 | c2pbzC_ |
|
not modelled |
99.1 |
15 |
PDB header:ligase
Chain: C: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of an imp biosynthesis protein purp from2 thermococcus kodakaraensis
|
| 80 | c1wr2A_ |
|
not modelled |
99.1 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein ph1789;
PDBTitle: crystal structure of ph1788 from pyrococcus horikoshii ot3
|
| 81 | d1a9xa3 |
|
not modelled |
99.1 |
22 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 82 | c2nu9E_ |
|
not modelled |
99.0 |
18 |
PDB header:ligase
Chain: E: PDB Molecule:succinyl-coa synthetase beta chain;
PDBTitle: c123at mutant of e. coli succinyl-coa synthetase2 orthorhombic crystal form
|
| 83 | d1kjqa1 |
|
not modelled |
99.0 |
100 |
Fold:Barrel-sandwich hybrid
Superfamily:Rudiment single hybrid motif
Family:BC C-terminal domain-like |
| 84 | d2j9ga2 |
|
not modelled |
99.0 |
21 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 85 | d1ulza2 |
|
not modelled |
98.9 |
19 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 86 | d1eucb2 |
|
not modelled |
98.7 |
17 |
Fold:ATP-grasp
Superfamily:Glutathione synthetase ATP-binding domain-like
Family:Succinyl-CoA synthetase, beta-chain, N-terminal domain |
| 87 | d2pbza2 |
|
not modelled |
98.7 |
14 |
Fold:ATP-grasp
Superfamily:Glutathione synthetase ATP-binding domain-like
Family:PurP ATP-binding domain-like |
| 88 | d2nu7b2 |
|
not modelled |
98.7 |
19 |
Fold:ATP-grasp
Superfamily:Glutathione synthetase ATP-binding domain-like
Family:Succinyl-CoA synthetase, beta-chain, N-terminal domain |
| 89 | c1eucB_ |
|
not modelled |
98.6 |
17 |
PDB header:ligase
Chain: B: PDB Molecule:succinyl-coa synthetase, beta chain;
PDBTitle: crystal structure of dephosphorylated pig heart, gtp-2 specific succinyl-coa synthetase
|
| 90 | d1w96a2 |
|
not modelled |
98.6 |
14 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 91 | d1gsoa2 |
|
not modelled |
98.3 |
24 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 92 | d3etja1 |
|
not modelled |
98.2 |
13 |
Fold:Barrel-sandwich hybrid
Superfamily:Rudiment single hybrid motif
Family:BC C-terminal domain-like |
| 93 | c3eywA_ |
|
not modelled |
98.0 |
15 |
PDB header:transport protein
Chain: A: PDB Molecule:c-terminal domain of glutathione-regulated potassium-efflux
PDBTitle: crystal structure of the c-terminal domain of e. coli kefc in complex2 with keff
|
| 94 | d1db3a_ |
|
not modelled |
97.9 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 95 | c1z45A_ |
|
not modelled |
97.9 |
19 |
PDB header:isomerase
Chain: A: PDB Molecule:gal10 bifunctional protein;
PDBTitle: crystal structure of the gal10 fusion protein galactose2 mutarotase/udp-galactose 4-epimerase from saccharomyces3 cerevisiae complexed with nad, udp-glucose, and galactose
|
| 96 | c2q1wC_ |
|
not modelled |
97.8 |
20 |
PDB header:sugar binding protein
Chain: C: PDB Molecule:putative nucleotide sugar epimerase/ dehydratase;
PDBTitle: crystal structure of the bordetella bronchiseptica enzyme wbmh in2 complex with nad+
|
| 97 | d1n7ha_ |
|
not modelled |
97.8 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 98 | c1e5lA_ |
|
not modelled |
97.7 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:saccharopine reductase;
PDBTitle: apo saccharopine reductase from magnaporthe grisea
|
| 99 | c3tinA_ |
|
not modelled |
97.7 |
21 |
PDB header:ligase
Chain: A: PDB Molecule:ttl protein;
PDBTitle: tubulin tyrosine ligase
|
| 100 | c2g1uA_ |
|
not modelled |
97.6 |
15 |
PDB header:transport protein
Chain: A: PDB Molecule:hypothetical protein tm1088a;
PDBTitle: crystal structure of a putative transport protein (tm1088a) from2 thermotoga maritima at 1.50 a resolution
|
| 101 | d1wvga1 |
|
not modelled |
97.6 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 102 | d1rkxa_ |
|
not modelled |
97.6 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 103 | c2pzlB_ |
|
not modelled |
97.6 |
20 |
PDB header:sugar binding protein
Chain: B: PDB Molecule:putative nucleotide sugar epimerase/ dehydratase;
PDBTitle: crystal structure of the bordetella bronchiseptica enzyme2 wbmg in complex with nad and udp
|
| 104 | c3mwdA_ |
|
not modelled |
97.6 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:atp-citrate synthase;
PDBTitle: truncated human atp-citrate lyase with citrate bound
|
| 105 | c1n7gB_ |
|
not modelled |
97.6 |
22 |
PDB header:lyase
Chain: B: PDB Molecule:gdp-d-mannose-4,6-dehydratase;
PDBTitle: crystal structure of the gdp-mannose 4,6-dehydratase2 ternary complex with nadph and gdp-rhamnose.
|
| 106 | d1lssa_ |
|
not modelled |
97.6 |
11 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Potassium channel NAD-binding domain |
| 107 | c1t2aC_ |
|
not modelled |
97.6 |
20 |
PDB header:structural genomics,lyase
Chain: C: PDB Molecule:gdp-mannose 4,6 dehydratase;
PDBTitle: crystal structure of human gdp-d-mannose 4,6-dehydratase
|
| 108 | d1t2aa_ |
|
not modelled |
97.6 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 109 | c3enkB_ |
|
not modelled |
97.6 |
18 |
PDB header:isomerase
Chain: B: PDB Molecule:udp-glucose 4-epimerase;
PDBTitle: 1.9a crystal structure of udp-glucose 4-epimerase from2 burkholderia pseudomallei
|
| 110 | d1rpna_ |
|
not modelled |
97.6 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 111 | c3ic5A_ |
|
not modelled |
97.5 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative saccharopine dehydrogenase;
PDBTitle: n-terminal domain of putative saccharopine dehydrogenase from ruegeria2 pomeroyi.
|
| 112 | c2pk3B_ |
|
not modelled |
97.5 |
26 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:gdp-6-deoxy-d-lyxo-4-hexulose reductase;
PDBTitle: crystal structure of a gdp-4-keto-6-deoxy-d-mannose reductase
|
| 113 | c2z1mC_ |
|
not modelled |
97.5 |
19 |
PDB header:lyase
Chain: C: PDB Molecule:gdp-d-mannose dehydratase;
PDBTitle: crystal structure of gdp-d-mannose dehydratase from aquifex aeolicus2 vf5
|
| 114 | d2c5aa1 |
|
not modelled |
97.5 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 115 | d1vl0a_ |
|
not modelled |
97.5 |
24 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 116 | c2p5uC_ |
|
not modelled |
97.5 |
20 |
PDB header:isomerase
Chain: C: PDB Molecule:udp-glucose 4-epimerase;
PDBTitle: crystal structure of thermus thermophilus hb8 udp-glucose 4-2 epimerase complex with nad
|
| 117 | c2x4gA_ |
|
not modelled |
97.5 |
21 |
PDB header:isomerase
Chain: A: PDB Molecule:nucleoside-diphosphate-sugar epimerase;
PDBTitle: crystal structure of pa4631, a nucleoside-diphosphate-sugar2 epimerase from pseudomonas aeruginosa
|
| 118 | d2jfga1 |
|
not modelled |
97.4 |
20 |
Fold:MurCD N-terminal domain
Superfamily:MurCD N-terminal domain
Family:MurCD N-terminal domain |
| 119 | c3sc6F_ |
|
not modelled |
97.4 |
25 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:dtdp-4-dehydrorhamnose reductase;
PDBTitle: 2.65 angstrom resolution crystal structure of dtdp-4-dehydrorhamnose2 reductase (rfbd) from bacillus anthracis str. ames in complex with3 nadp
|
| 120 | d1pjca1 |
|
not modelled |
97.4 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |

