1 c1ezaA_
100.0
28
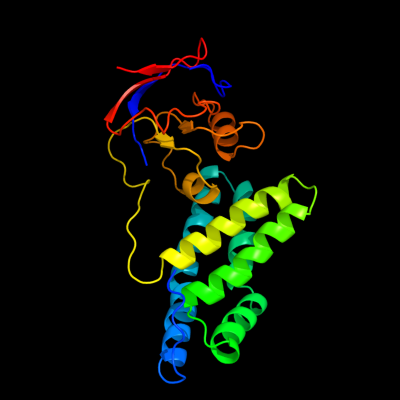
PDB header: phosphotransferaseChain: A: PDB Molecule: enzyme i;PDBTitle: amino terminal domain of enzyme i from escherichia coli nmr,2 restrained regularized mean structure
2 c2hroA_
100.0
24
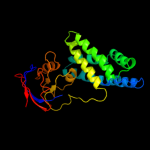
PDB header: transferaseChain: A: PDB Molecule: phosphoenolpyruvate-protein phosphotransferase;PDBTitle: structure of the full-lenght enzyme i of the pts system from2 staphylococcus carnosus
3 c2hwgA_
100.0
28
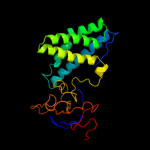
PDB header: transferaseChain: A: PDB Molecule: phosphoenolpyruvate-protein phosphotransferase;PDBTitle: structure of phosphorylated enzyme i of the2 phosphoenolpyruvate:sugar phosphotransferase system
4 d3ct6a1
100.0
30
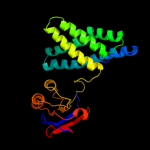
Fold: PTS system fructose IIA component-likeSuperfamily: PTS system fructose IIA component-likeFamily: DhaM-like5 d1zyma1
99.9
30
Fold: SAM domain-likeSuperfamily: Enzyme I of the PEP:sugar phosphotransferase system HPr-binding (sub)domainFamily: Enzyme I of the PEP:sugar phosphotransferase system HPr-binding (sub)domain6 d3b48a1
99.9
30
Fold: PTS system fructose IIA component-likeSuperfamily: PTS system fructose IIA component-likeFamily: DhaM-like7 d2hpra_
99.9
31
Fold: HPr-likeSuperfamily: HPr-likeFamily: HPr-like8 c3gdwA_
99.8
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: sigma-54 interaction domain protein;PDBTitle: crystal structure of sigma-54 interaction domain protein from2 enterococcus faecalis
9 d1ka5a_
99.8
31
Fold: HPr-likeSuperfamily: HPr-likeFamily: HPr-like10 d1pcha_
99.8
24
Fold: HPr-likeSuperfamily: HPr-likeFamily: HPr-like11 c3le1B_
99.8
38
PDB header: transferaseChain: B: PDB Molecule: phosphotransferase system, hpr-related proteins;PDBTitle: crystal structure of apohpr monomer from thermoanaerobacter2 tengcongensis
12 d1qr5a_
99.8
31
Fold: HPr-likeSuperfamily: HPr-likeFamily: HPr-like13 d1mo1a_
99.8
31
Fold: HPr-likeSuperfamily: HPr-likeFamily: HPr-like14 c3ihsB_
99.8
32
PDB header: transport proteinChain: B: PDB Molecule: phosphocarrier protein hpr;PDBTitle: crystal structure of a phosphocarrier protein hpr from2 bacillus anthracis str. ames
15 d1zyma2
99.8
26
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: Phosphohistidine domainFamily: N-terminal domain of enzyme I of the PEP:sugar phosphotransferase system16 d2nzul1
99.8
31
Fold: HPr-likeSuperfamily: HPr-likeFamily: HPr-like17 d1ptfa_
99.8
30
Fold: HPr-likeSuperfamily: HPr-likeFamily: HPr-like18 d1cm3a_
99.8
23
Fold: HPr-likeSuperfamily: HPr-likeFamily: HPr-like19 d1zvvj1
99.8
29
Fold: HPr-likeSuperfamily: HPr-likeFamily: HPr-like20 c1h6zA_
99.7
21
PDB header: transferaseChain: A: PDB Molecule: pyruvate phosphate dikinase;PDBTitle: 3.0 a resolution crystal structure of glycosomal pyruvate2 phosphate dikinase from trypanosoma brucei
21 c1kblA_
not modelled
99.7
25
PDB header: transferaseChain: A: PDB Molecule: pyruvate phosphate dikinase;PDBTitle: pyruvate phosphate dikinase
22 d1h6za2
not modelled
99.7
22
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: Phosphohistidine domainFamily: Pyruvate phosphate dikinase, central domain23 d1kbla2
not modelled
99.7
28
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: Phosphohistidine domainFamily: Pyruvate phosphate dikinase, central domain24 d1vbga2
not modelled
99.7
26
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: Phosphohistidine domainFamily: Pyruvate phosphate dikinase, central domain25 c1vbhA_
not modelled
99.5
25
PDB header: transferaseChain: A: PDB Molecule: pyruvate,orthophosphate dikinase;PDBTitle: pyruvate phosphate dikinase with bound mg-pep from maize
26 c3gx1A_
not modelled
99.5
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: lin1832 protein;PDBTitle: crystal structure of a domain of lin1832 from listeria innocua
27 c2olsA_
not modelled
99.5
18
PDB header: transferaseChain: A: PDB Molecule: phosphoenolpyruvate synthase;PDBTitle: the crystal structure of the phosphoenolpyruvate synthase from2 neisseria meningitidis
28 c3iprC_
not modelled
99.4
23
PDB header: transferaseChain: C: PDB Molecule: pts system, iia component;PDBTitle: crystal structure of the enterococcus faecalis gluconate2 specific eiia phosphotransferase system component
29 c3t07D_
not modelled
99.2
19
PDB header: transferase/transferase inhibitorChain: D: PDB Molecule: pyruvate kinase;PDBTitle: crystal structure of s. aureus pyruvate kinase in complex with a2 naturally occurring bis-indole alkaloid
30 c2e28A_
not modelled
99.0
19
PDB header: transferaseChain: A: PDB Molecule: pyruvate kinase;PDBTitle: crystal structure analysis of pyruvate kinase from bacillus2 stearothermophilus
31 d1pdoa_
not modelled
98.5
14
Fold: PTS system fructose IIA component-likeSuperfamily: PTS system fructose IIA component-likeFamily: EIIA-man component-like32 c3mtqA_
not modelled
97.9
16
PDB header: transferaseChain: A: PDB Molecule: putative phosphoenolpyruvate-dependent sugarPDBTitle: crystal structure of a putative phosphoenolpyruvate-dependent sugar2 phosphotransferase system (pts) permease (kpn_04802) from klebsiella3 pneumoniae subsp. pneumoniae mgh 78578 at 1.70 a resolution
33 c3lfhF_
not modelled
97.1
14
PDB header: transferaseChain: F: PDB Molecule: phosphotransferase system, mannose/fructose-specificPDBTitle: crystal structure of manxa from thermoanaerobacter tengcongensis
34 d3beda1
not modelled
97.1
14
Fold: PTS system fructose IIA component-likeSuperfamily: PTS system fructose IIA component-likeFamily: EIIA-man component-like35 d1un8a4
96.9
18
Fold: DAK1/DegV-likeSuperfamily: DAK1/DegV-likeFamily: DAK136 c3ct4B_
not modelled
96.2
22
PDB header: transferaseChain: B: PDB Molecule: pts-dependent dihydroxyacetone kinase,PDBTitle: structure of dha-kinase subunit dhak from l. lactis
37 c1un9B_
not modelled
96.2
18
PDB header: kinaseChain: B: PDB Molecule: dihydroxyacetone kinase;PDBTitle: crystal structure of the dihydroxyacetone kinase from c.2 freundii in complex with amp-pnp and mg2+
38 d1oi2a_
not modelled
96.2
23
Fold: DAK1/DegV-likeSuperfamily: DAK1/DegV-likeFamily: DAK139 c2iu6B_
not modelled
95.7
18
PDB header: transferaseChain: B: PDB Molecule: dihydroxyacetone kinase;PDBTitle: regulation of the dha operon of lactococcus lactis
40 d2hi6a1
not modelled
87.1
18
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: LeuD/IlvD-likeFamily: AF0055-like41 c3nyiA_
not modelled
84.0
11
PDB header: lipid binding proteinChain: A: PDB Molecule: fat acid-binding protein;PDBTitle: the crystal structure of a fat acid (stearic acid)-binding protein2 from eubacterium ventriosum atcc 27560.
42 d1eepa_
not modelled
78.5
11
Fold: TIM beta/alpha-barrelSuperfamily: Inosine monophosphate dehydrogenase (IMPDH)Family: Inosine monophosphate dehydrogenase (IMPDH)43 c3navB_
not modelled
76.1
25
PDB header: lyaseChain: B: PDB Molecule: tryptophan synthase alpha chain;PDBTitle: crystal structure of an alpha subunit of tryptophan synthase from2 vibrio cholerae o1 biovar el tor str. n16961
44 c2a7rD_
not modelled
76.0
15
PDB header: oxidoreductaseChain: D: PDB Molecule: gmp reductase 2;PDBTitle: crystal structure of human guanosine monophosphate2 reductase 2 (gmpr2)
45 c2px0D_
not modelled
70.0
13
PDB header: biosynthetic proteinChain: D: PDB Molecule: flagellar biosynthesis protein flhf;PDBTitle: crystal structure of flhf complexed with gmppnp/mg(2+)
46 d1ivsa1
not modelled
69.3
15
Fold: Long alpha-hairpinSuperfamily: tRNA-binding armFamily: Valyl-tRNA synthetase (ValRS) C-terminal domain47 c3jr7A_
not modelled
62.9
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized egv family protein cog1307;PDBTitle: the crystal structure of the protein of degv family cog1307 with2 unknown function from ruminococcus gnavus atcc 29149
48 c1mgpA_
not modelled
62.0
21
PDB header: lipid binding proteinChain: A: PDB Molecule: hypothetical protein tm841;PDBTitle: hypothetical protein tm841 from thermotoga maritima reveals2 fatty acid binding function
49 d1mgpa_
not modelled
62.0
21
Fold: DAK1/DegV-likeSuperfamily: DAK1/DegV-likeFamily: DegV-like50 d2auna1
not modelled
61.3
18
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: LD-carboxypeptidase A C-terminal domain-likeFamily: LD-carboxypeptidase A C-terminal domain-like51 c3thaB_
not modelled
59.8
17
PDB header: lyaseChain: B: PDB Molecule: tryptophan synthase alpha chain;PDBTitle: tryptophan synthase subunit alpha from campylobacter jejuni.
52 d1vrda1
not modelled
57.4
11
Fold: TIM beta/alpha-barrelSuperfamily: Inosine monophosphate dehydrogenase (IMPDH)Family: Inosine monophosphate dehydrogenase (IMPDH)53 d2gp4a1
not modelled
57.0
11
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: LeuD/IlvD-likeFamily: IlvD/EDD C-terminal domain-like54 c1nw3A_
not modelled
56.7
28
PDB header: transferaseChain: A: PDB Molecule: histone methyltransferase dot1l;PDBTitle: structure of the catalytic domain of human dot1l, a non-set2 domain nucleosomal histone methyltransferase
55 d1nw3a_
not modelled
56.7
28
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Catalytic, N-terminal domain of histone methyltransferase Dot1l56 d1jr1a1
not modelled
56.3
9
Fold: TIM beta/alpha-barrelSuperfamily: Inosine monophosphate dehydrogenase (IMPDH)Family: Inosine monophosphate dehydrogenase (IMPDH)57 c2g7zB_
not modelled
56.0
13
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: conserved hypothetical protein spy1493;PDBTitle: conserved degv-like protein of unknown function from streptococcus2 pyogenes m1 gas binds long-chain fatty acids
58 d1oi7a2
not modelled
54.9
18
Fold: Flavodoxin-likeSuperfamily: Succinyl-CoA synthetase domainsFamily: Succinyl-CoA synthetase domains59 c1u2zC_
not modelled
54.9
18
PDB header: transferaseChain: C: PDB Molecule: histone-lysine n-methyltransferase, h3 lysine-79PDBTitle: crystal structure of histone k79 methyltransferase dot1p2 from yeast
60 c2dt8A_
not modelled
54.2
21
PDB header: lipid binding proteinChain: A: PDB Molecule: degv family protein;PDBTitle: fatty acid binding of a degv family protein from thermus thermophilus
61 c2jgqB_
not modelled
53.9
18
PDB header: isomeraseChain: B: PDB Molecule: triosephosphate isomerase;PDBTitle: kinetics and structural properties of triosephosphate2 isomerase from helicobacter pylori
62 c3iz5H_
not modelled
52.6
24
PDB header: ribosomeChain: H: PDB Molecule: 60s ribosomal protein l7a (l7ae);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
63 c1vrdA_
not modelled
52.0
11
PDB header: oxidoreductaseChain: A: PDB Molecule: inosine-5'-monophosphate dehydrogenase;PDBTitle: crystal structure of inosine-5'-monophosphate dehydrogenase (tm1347)2 from thermotoga maritima at 2.18 a resolution
64 c1w2wJ_
not modelled
50.6
19
PDB header: isomeraseChain: J: PDB Molecule: 5-methylthioribose-1-phosphate isomerase;PDBTitle: crystal structure of yeast ypr118w, a methylthioribose-1-2 phosphate isomerase related to regulatory eif2b subunits
65 c4a1eF_
not modelled
48.0
24
PDB header: ribosomeChain: F: PDB Molecule: rpl7a;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna, 5.8s rrna3 and proteins of molecule 1
66 d1u2za_
not modelled
47.8
18
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Catalytic, N-terminal domain of histone methyltransferase Dot1l67 d1zfja1
not modelled
47.7
13
Fold: TIM beta/alpha-barrelSuperfamily: Inosine monophosphate dehydrogenase (IMPDH)Family: Inosine monophosphate dehydrogenase (IMPDH)68 d2fzva1
not modelled
47.6
15
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: NADPH-dependent FMN reductase69 c3pl5A_
not modelled
46.5
14
PDB header: lipid binding proteinChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: fatty acid binding protein
70 d2nu7a2
not modelled
45.7
21
Fold: Flavodoxin-likeSuperfamily: Succinyl-CoA synthetase domainsFamily: Succinyl-CoA synthetase domains71 c2fzvC_
not modelled
44.6
16
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: putative arsenical resistance protein;PDBTitle: crystal structure of an apo form of a flavin-binding protein from2 shigella flexneri
72 d1qopa_
not modelled
44.4
19
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes73 c2zkrf_
not modelled
43.8
28
PDB header: ribosomal protein/rnaChain: F: PDB Molecule: rna expansion segment es7 part iii;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
74 c3khjE_
not modelled
43.8
13
PDB header: oxidoreductaseChain: E: PDB Molecule: inosine-5-monophosphate dehydrogenase;PDBTitle: c. parvum inosine monophosphate dehydrogenase bound by inhibitor c64
75 c2yvkA_
not modelled
43.4
15
PDB header: isomeraseChain: A: PDB Molecule: methylthioribose-1-phosphate isomerase;PDBTitle: crystal structure of 5-methylthioribose 1-phosphate2 isomerase product complex from bacillus subtilis
76 c2ekcA_
not modelled
43.3
24
PDB header: lyaseChain: A: PDB Molecule: tryptophan synthase alpha chain;PDBTitle: structural study of project id aq_1548 from aquifex aeolicus vf5
77 c3izcH_
not modelled
43.2
26
PDB header: ribosomeChain: H: PDB Molecule: 60s ribosomal protein rpl8 (l7ae);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
78 c2gp4A_
not modelled
41.5
11
PDB header: lyaseChain: A: PDB Molecule: 6-phosphogluconate dehydratase;PDBTitle: structure of [fes]cluster-free apo form of 6-phosphogluconate2 dehydratase from shewanella oneidensis
79 d1cf7b_
not modelled
39.8
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Cell cycle transcription factor e2f-dp80 c3r2gA_
not modelled
38.9
13
PDB header: oxidoreductaseChain: A: PDB Molecule: inosine 5'-monophosphate dehydrogenase;PDBTitle: crystal structure of inosine 5' monophosphate dehydrogenase from2 legionella pneumophila
81 d1pzxa_
not modelled
38.1
10
Fold: DAK1/DegV-likeSuperfamily: DAK1/DegV-likeFamily: DegV-like82 d1vzwa1
not modelled
36.6
23
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Histidine biosynthesis enzymes83 c2jpiA_
not modelled
34.5
13
PDB header: structural genomicsChain: A: PDB Molecule: hypothetical protein;PDBTitle: chemical shift assignments of pa4090 from pseudomonas2 aeruginosa
84 d1y0ea_
not modelled
34.1
12
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: NanE-like85 d1g0da4
not modelled
33.2
29
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Transglutaminase core86 d1tuba1
not modelled
33.2
13
Fold: Tubulin nucleotide-binding domain-likeSuperfamily: Tubulin nucleotide-binding domain-likeFamily: Tubulin, GTPase domain87 d1piia2
not modelled
33.1
29
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes88 d1j5ta_
not modelled
32.1
28
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes89 c1jcnA_
not modelled
32.0
10
PDB header: oxidoreductaseChain: A: PDB Molecule: inosine monophosphate dehydrogenase i;PDBTitle: binary complex of human type-i inosine monophosphate dehydrogenase2 with 6-cl-imp
90 d1hg3a_
not modelled
31.7
27
Fold: TIM beta/alpha-barrelSuperfamily: Triosephosphate isomerase (TIM)Family: Triosephosphate isomerase (TIM)91 d2csua2
not modelled
31.7
19
Fold: Flavodoxin-likeSuperfamily: Succinyl-CoA synthetase domainsFamily: Succinyl-CoA synthetase domains92 c3dmaA_
not modelled
31.5
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: exopolyphosphatase-related protein;PDBTitle: crystal structure of an exopolyphosphatase-related protein2 from bacteroides fragilis. northeast structural genomics3 target bfr192
93 d1tb3a1
not modelled
31.3
18
Fold: TIM beta/alpha-barrelSuperfamily: FMN-linked oxidoreductasesFamily: FMN-linked oxidoreductases94 c1oi7A_
not modelled
30.1
18
PDB header: synthetaseChain: A: PDB Molecule: succinyl-coa synthetase alpha chain;PDBTitle: the crystal structure of succinyl-coa synthetase alpha2 subunit from thermus thermophilus
95 d2btma_
not modelled
30.0
19
Fold: TIM beta/alpha-barrelSuperfamily: Triosephosphate isomerase (TIM)Family: Triosephosphate isomerase (TIM)96 c2fpgA_
not modelled
29.1
21
PDB header: ligaseChain: A: PDB Molecule: succinyl-coa ligase [gdp-forming] alpha-chain,PDBTitle: crystal structure of pig gtp-specific succinyl-coa2 synthetase in complex with gdp
97 d1pvna1
not modelled
27.5
11
Fold: TIM beta/alpha-barrelSuperfamily: Inosine monophosphate dehydrogenase (IMPDH)Family: Inosine monophosphate dehydrogenase (IMPDH)98 c3lupA_
not modelled
27.1
10
PDB header: structure genomics, unknown functionChain: A: PDB Molecule: degv family protein;PDBTitle: crystal structure of fatty acid binding degv family protein sag13422 from streptococcus agalactiae
99 d1wv2a_
not modelled
26.5
29
Fold: TIM beta/alpha-barrelSuperfamily: ThiG-likeFamily: ThiG-like100 d1o8bb1
not modelled
25.7
29
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: D-ribose-5-phosphate isomerase (RpiA), catalytic domain101 d1v7la_
not modelled
25.6
21
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: LeuD/IlvD-likeFamily: LeuD-like102 d1aopa2
not modelled
25.4
18
Fold: Ferredoxin-likeSuperfamily: Nitrite/Sulfite reductase N-terminal domain-likeFamily: Duplicated SiR/NiR-like domains 1 and 3103 c2rkbE_
not modelled
25.1
25
PDB header: lyaseChain: E: PDB Molecule: serine dehydratase-like;PDBTitle: serine dehydratase like-1 from human cancer cells
104 d1a53a_
not modelled
24.8
18
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes105 d1i4na_
not modelled
24.8
26
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes106 d2cu0a1
not modelled
24.6
14
Fold: TIM beta/alpha-barrelSuperfamily: Inosine monophosphate dehydrogenase (IMPDH)Family: Inosine monophosphate dehydrogenase (IMPDH)107 c2k8hA_
not modelled
24.3
16
PDB header: signaling proteinChain: A: PDB Molecule: small ubiquitin protein;PDBTitle: solution structure of sumo from trypanosoma brucei
108 c2nu8D_
not modelled
23.9
20
PDB header: ligaseChain: D: PDB Molecule: succinyl-coa ligase [adp-forming] subunit alpha;PDBTitle: c123at mutant of e. coli succinyl-coa synthetase
109 c1ypfB_
not modelled
23.7
14
PDB header: oxidoreductaseChain: B: PDB Molecule: gmp reductase;PDBTitle: crystal structure of guac (ba5705) from bacillus anthracis at 1.8 a2 resolution
110 c3fdjA_
not modelled
23.6
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: degv family protein;PDBTitle: the structure of a degv family protein from eubacterium eligens.
111 c2ekeC_
not modelled
23.6
8
PDB header: ligase/protein bindingChain: C: PDB Molecule: ubiquitin-like protein smt3;PDBTitle: structure of a sumo-binding-motif mimic bound to smt3p-2 ubc9p: conservation of a noncovalent ubiquitin-like3 protein-e2 complex as a platform for selective4 interactions within a sumo pathway
112 d1geqa_
not modelled
23.5
28
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes113 c3no4A_
not modelled
23.1
25
PDB header: hydrolaseChain: A: PDB Molecule: creatinine amidohydrolase;PDBTitle: crystal structure of a creatinine amidohydrolase (npun_f1913) from2 nostoc punctiforme pcc 73102 at 2.00 a resolution
114 c2dy0A_
not modelled
23.0
20
PDB header: transferaseChain: A: PDB Molecule: adenine phosphoribosyltransferase;PDBTitle: crystal structure of project jw0458 from escherichia coli
115 c2rbfB_
not modelled
22.8
13
PDB header: oxidoreductase/dnaChain: B: PDB Molecule: bifunctional protein puta;PDBTitle: structure of the ribbon-helix-helix domain of escherichia coli puta2 (puta52) complexed with operator dna (o2)
116 d1ka9f_
not modelled
22.6
24
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Histidine biosynthesis enzymes117 d2pw9a1
not modelled
22.4
13
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: FdhD/NarQ118 c3m3hA_
not modelled
22.2
10
PDB header: transferaseChain: A: PDB Molecule: orotate phosphoribosyltransferase;PDBTitle: 1.75 angstrom resolution crystal structure of an orotate2 phosphoribosyltransferase from bacillus anthracis str. 'ames3 ancestor'
119 d1h41a1
not modelled
22.1
18
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: alpha-D-glucuronidase/Hyaluronidase catalytic domain120 c3edlA_
not modelled
21.8
15
PDB header: structural proteinChain: A: PDB Molecule: alpha-tubulin;PDBTitle: kinesin13-microtubule ring complex