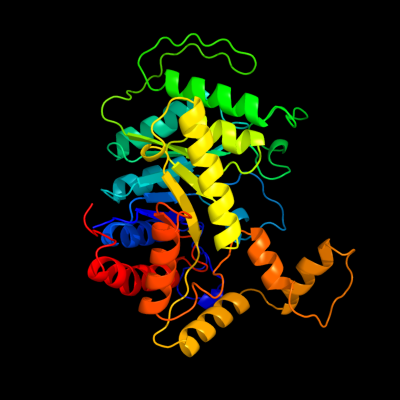
| 1 | c3sdoB_
|
|
 |
100.0 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nitrilotriacetate monooxygenase;
PDBTitle: structure of a nitrilotriacetate monooxygenase from burkholderia2 pseudomallei
|
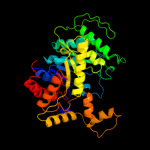
| 2 | c1tvlA_
|
|
 |
100.0 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein ytnj;
PDBTitle: structure of ytnj from bacillus subtilis
|
| 3 | d1tvla_
|
|
 |
100.0 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Ssud-like monoxygenases |
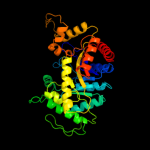
| 4 | c3raoB_
|
|
 |
100.0 |
25 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative luciferase-like monooxygenase;
PDBTitle: crystal structure of the luciferase-like monooxygenase from bacillus2 cereus atcc 10987.
|
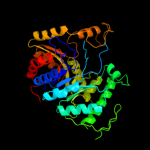
| 5 | c3b9nB_
|
|
 |
100.0 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:alkane monoxygenase;
PDBTitle: crystal structure of long-chain alkane monooxygenase (lada)
|
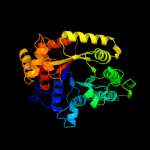
| 6 | d1nqka_
|
|
 |
100.0 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Ssud-like monoxygenases |
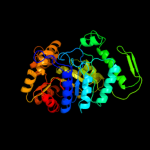
| 7 | c2wgkA_
|
|
 |
100.0 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3,6-diketocamphane 1,6 monooxygenase;
PDBTitle: type ii baeyer-villiger monooxygenase oxygenating2 constituent of 3,6-diketocamphane 1,6 monooxygenase from3 pseudomonas putida
|
| 8 | d1luca_
|
|
 |
100.0 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Bacterial luciferase (alkanal monooxygenase) |
| 9 | d1lucb_
|
|
 |
100.0 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Bacterial luciferase (alkanal monooxygenase) |
| 10 | c1z69D_
|
|
 |
100.0 |
17 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:coenzyme f420-dependent n(5),n(10)-
PDBTitle: crystal structure of methylenetetrahydromethanopterin2 reductase (mer) in complex with coenzyme f420
|
| 11 | d1ezwa_
|
|
 |
100.0 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:F420 dependent oxidoreductases |
| 12 | c2i7gA_
|
|
 |
100.0 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:monooxygenase;
PDBTitle: crystal structure of monooxygenase from agrobacterium tumefaciens
|
| 13 | c3c8nB_
|
|
 |
100.0 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable f420-dependent glucose-6-phosphate dehydrogenase
PDBTitle: crystal structure of apo-fgd1 from mycobacterium tuberculosis
|
| 14 | d1f07a_
|
|
 |
100.0 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:F420 dependent oxidoreductases |
| 15 | d1rhca_
|
|
 |
100.0 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:F420 dependent oxidoreductases |
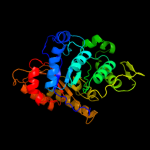
| 16 | c2b81D_
|
|
 |
100.0 |
17 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:luciferase-like monooxygenase;
PDBTitle: crystal structure of the luciferase-like monooxygenase from bacillus2 cereus
|
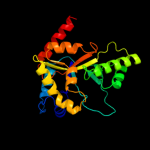
| 17 | d1nfpa_
|
|
 |
99.9 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Non-fluorescent flavoprotein (luxF, FP390) |
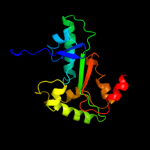
| 18 | d1fvpa_
|
|
 |
99.5 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Non-fluorescent flavoprotein (luxF, FP390) |
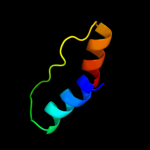
| 19 | c1zlpA_
|
|
 |
83.4 |
8 |
PDB header:lyase
Chain: A: PDB Molecule:petal death protein;
PDBTitle: petal death protein psr132 with cysteine-linked glutaraldehyde forming2 a thiohemiacetal adduct
|
| 20 | c3b8iF_
|
|
 |
82.5 |
19 |
PDB header:lyase
Chain: F: PDB Molecule:pa4872 oxaloacetate decarboxylase;
PDBTitle: crystal structure of oxaloacetate decarboxylase from pseudomonas2 aeruginosa (pa4872) in complex with oxalate and mg2+.
|
| 21 | c3fa4D_ |
|
not modelled |
82.1 |
11 |
PDB header:lyase
Chain: D: PDB Molecule:2,3-dimethylmalate lyase;
PDBTitle: crystal structure of 2,3-dimethylmalate lyase, a pep mutase/isocitrate2 lyase superfamily member, triclinic crystal form
|
| 22 | c3eooL_ |
|
not modelled |
81.9 |
14 |
PDB header:lyase
Chain: L: PDB Molecule:methylisocitrate lyase;
PDBTitle: 2.9a crystal structure of methyl-isocitrate lyase from2 burkholderia pseudomallei
|
| 23 | c2ze3A_ |
|
not modelled |
81.1 |
19 |
PDB header:isomerase
Chain: A: PDB Molecule:dfa0005;
PDBTitle: crystal structure of dfa0005 complexed with alpha-ketoglutarate: a2 novel member of the icl/pepm superfamily from alkali-tolerant3 deinococcus ficus
|
| 24 | c3lyeA_ |
|
not modelled |
81.1 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:oxaloacetate acetyl hydrolase;
PDBTitle: crystal structure of oxaloacetate acetylhydrolase
|
| 25 | d1ujqa_ |
|
not modelled |
80.7 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 26 | d1muma_ |
|
not modelled |
80.3 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 27 | c2qiwA_ |
|
not modelled |
79.4 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:pep phosphonomutase;
PDBTitle: crystal structure of a putative phosphoenolpyruvate phosphonomutase2 (ncgl1015, cgl1060) from corynebacterium glutamicum atcc 13032 at3 1.80 a resolution
|
| 28 | c3qy6A_ |
|
not modelled |
78.3 |
8 |
PDB header:hydrolase
Chain: A: PDB Molecule:tyrosine-protein phosphatase ywqe;
PDBTitle: crystal structures of ywqe from bacillus subtilis and cpsb from2 streptococcus pneumoniae, unique metal-dependent tyrosine3 phosphatases
|
| 29 | c3ih1A_ |
|
not modelled |
77.7 |
8 |
PDB header:lyase
Chain: A: PDB Molecule:methylisocitrate lyase;
PDBTitle: crystal structure of carboxyvinyl-carboxyphosphonate phosphorylmutase2 from bacillus anthracis
|
| 30 | c2x7vA_ |
|
not modelled |
68.2 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:probable endonuclease 4;
PDBTitle: crystal structure of thermotoga maritima endonuclease iv in2 the presence of zinc
|
| 31 | d1s2wa_ |
|
not modelled |
49.4 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 32 | c2wjeA_ |
|
not modelled |
49.2 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:tyrosine-protein phosphatase cpsb;
PDBTitle: crystal structure of the tyrosine phosphatase cps4b from2 steptococcus pneumoniae tigr4.
|
| 33 | d1hl9a2 |
|
not modelled |
43.8 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Putative alpha-L-fucosidase, catalytic domain |
| 34 | c2hjpA_ |
|
not modelled |
42.7 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:phosphonopyruvate hydrolase;
PDBTitle: crystal structure of phosphonopyruvate hydrolase complex with2 phosphonopyruvate and mg++
|
| 35 | c3ez4B_ |
|
not modelled |
36.8 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:3-methyl-2-oxobutanoate hydroxymethyltransferase;
PDBTitle: crystal structure of 3-methyl-2-oxobutanoate2 hydroxymethyltransferase from burkholderia pseudomallei
|
| 36 | d1jpdx1 |
|
not modelled |
35.1 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
| 37 | c3kwsB_ |
|
not modelled |
34.2 |
9 |
PDB header:isomerase
Chain: B: PDB Molecule:putative sugar isomerase;
PDBTitle: crystal structure of putative sugar isomerase (yp_001305149.1) from2 parabacteroides distasonis atcc 8503 at 1.68 a resolution
|
| 38 | d1r3sa_ |
|
not modelled |
29.6 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:UROD/MetE-like
Family:Uroporphyrinogen decarboxylase, UROD |
| 39 | c1jpkA_ |
|
not modelled |
28.6 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: gly156asp mutant of human urod, human uroporphyrinogen iii2 decarboxylase
|
| 40 | d2noca1 |
|
not modelled |
26.5 |
29 |
Fold:Dodecin subunit-like
Superfamily:YdgH-like
Family:YdgH-like |
| 41 | c1gshA_ |
|
not modelled |
26.3 |
16 |
PDB header:glutathione biosynthesis ligase
Chain: A: PDB Molecule:glutathione biosynthetic ligase;
PDBTitle: structure of escherichia coli glutathione synthetase at ph 7.5
|
| 42 | d2d69a1 |
|
not modelled |
26.2 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
| 43 | c3e02A_ |
|
not modelled |
26.2 |
25 |
PDB header:metal binding protein
Chain: A: PDB Molecule:uncharacterized protein duf849;
PDBTitle: crystal structure of a duf849 family protein (bxe_c0271) from2 burkholderia xenovorans lb400 at 1.90 a resolution
|
| 44 | c2hk1D_ |
|
not modelled |
25.8 |
22 |
PDB header:isomerase
Chain: D: PDB Molecule:d-psicose 3-epimerase;
PDBTitle: crystal structure of d-psicose 3-epimerase (dpease) in the presence of2 d-fructose
|
| 45 | d1xp3a1 |
|
not modelled |
25.3 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Endonuclease IV |
| 46 | c2r8wB_ |
|
not modelled |
24.8 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:agr_c_1641p;
PDBTitle: the crystal structure of dihydrodipicolinate synthase (atu0899) from2 agrobacterium tumefaciens str. c58
|
| 47 | d1j93a_ |
|
not modelled |
24.7 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:UROD/MetE-like
Family:Uroporphyrinogen decarboxylase, UROD |
| 48 | c3gzaB_ |
|
not modelled |
24.0 |
20 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative alpha-l-fucosidase;
PDBTitle: crystal structure of putative alpha-l-fucosidase (np_812709.1) from2 bacteroides thetaiotaomicron vpi-5482 at 1.60 a resolution
|
| 49 | c3bh1A_ |
|
not modelled |
22.4 |
32 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0371 protein dip2346;
PDBTitle: crystal structure of protein dip2346 from corynebacterium diphtheriae
|
| 50 | d1xkya1 |
|
not modelled |
21.9 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 51 | c1hl8B_ |
|
not modelled |
21.7 |
26 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative alpha-l-fucosidase;
PDBTitle: crystal structure of thermotoga maritima alpha-fucosidase
|
| 52 | c3bi8A_ |
|
not modelled |
20.8 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: structure of dihydrodipicolinate synthase from clostridium2 botulinum
|
| 53 | c2wvsD_ |
|
not modelled |
20.6 |
19 |
PDB header:hydrolase
Chain: D: PDB Molecule:alpha-l-fucosidase;
PDBTitle: crystal structure of an alpha-l-fucosidase gh29 trapped2 covalent intermediate from bacteroides thetaiotaomicron in3 complex with 2-fluoro-fucosyl fluoride using an e288q4 mutant
|
| 54 | c3pueA_ |
|
not modelled |
20.0 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of the complex of dhydrodipicolinate synthase from2 acinetobacter baumannii with lysine at 2.6a resolution
|
| 55 | c3e38A_ |
|
not modelled |
19.4 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:two-domain protein containing predicted php-like metal-
PDBTitle: crystal structure of a two-domain protein containing predicted php-2 like metal-dependent phosphoesterase (bvu_3505) from bacteroides3 vulgatus atcc 8482 at 2.20 a resolution
|
| 56 | c2nuxB_ |
|
not modelled |
18.1 |
8 |
PDB header:lyase
Chain: B: PDB Molecule:2-keto-3-deoxygluconate/2-keto-3-deoxy-6-phospho gluconate
PDBTitle: 2-keto-3-deoxygluconate aldolase from sulfolobus acidocaldarius,2 native structure in p6522 at 2.5 a resolution
|
| 57 | d2a6na1 |
|
not modelled |
17.1 |
7 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 58 | c1m7xC_ |
|
not modelled |
16.8 |
27 |
PDB header:transferase
Chain: C: PDB Molecule:1,4-alpha-glucan branching enzyme;
PDBTitle: the x-ray crystallographic structure of branching enzyme
|
| 59 | c3lerA_ |
|
not modelled |
16.1 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 campylobacter jejuni subsp. jejuni nctc 11168
|
| 60 | c3k1dA_ |
|
not modelled |
16.0 |
31 |
PDB header:transferase
Chain: A: PDB Molecule:1,4-alpha-glucan-branching enzyme;
PDBTitle: crystal structure of glycogen branching enzyme synonym: 1,4-alpha-d-2 glucan:1,4-alpha-d-glucan 6-glucosyl-transferase from mycobacterium3 tuberculosis h37rv
|
| 61 | d1oyaa_ |
|
not modelled |
15.2 |
4 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 62 | c2r94B_ |
|
not modelled |
15.1 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:2-keto-3-deoxy-(6-phospho-)gluconate aldolase;
PDBTitle: crystal structure of kd(p)ga from t.tenax
|
| 63 | d1gxha_ |
|
not modelled |
15.0 |
27 |
Fold:Acyl carrier protein-like
Superfamily:Colicin E immunity proteins
Family:Colicin E immunity proteins |
| 64 | d8ruca1 |
|
not modelled |
14.0 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
| 65 | c2ejaB_ |
|
not modelled |
13.9 |
10 |
PDB header:lyase
Chain: B: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: crystal structure of uroporphyrinogen decarboxylase from2 aquifex aeolicus
|
| 66 | d1z41a1 |
|
not modelled |
13.5 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 67 | c3gg2B_ |
|
not modelled |
13.4 |
9 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:sugar dehydrogenase, udp-glucose/gdp-mannose
PDBTitle: crystal structure of udp-glucose 6-dehydrogenase from2 porphyromonas gingivalis bound to product udp-glucuronate
|
| 68 | c2hmcA_ |
|
not modelled |
13.3 |
9 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: the crystal structure of dihydrodipicolinate synthase dapa from2 agrobacterium tumefaciens
|
| 69 | d1m6ya2 |
|
not modelled |
13.2 |
11 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:MraW-like putative methyltransferases |
| 70 | c3dcpB_ |
|
not modelled |
12.9 |
24 |
PDB header:hydrolase
Chain: B: PDB Molecule:histidinol-phosphatase;
PDBTitle: crystal structure of the putative histidinol phosphatase2 hisk from listeria monocytogenes. northeast structural3 genomics consortium target lmr141.
|
| 71 | c2infB_ |
|
not modelled |
12.8 |
10 |
PDB header:lyase
Chain: B: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: crystal structure of uroporphyrinogen decarboxylase from2 bacillus subtilis
|
| 72 | c3na8A_ |
|
not modelled |
12.4 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:putative dihydrodipicolinate synthetase;
PDBTitle: crystal structure of a putative dihydrodipicolinate synthetase from2 pseudomonas aeruginosa
|
| 73 | c1rcxH_ |
|
not modelled |
12.0 |
10 |
PDB header:lyase (carbon-carbon)
Chain: H: PDB Molecule:ribulose bisphosphate carboxylase/oxygenase;
PDBTitle: non-activated spinach rubisco in complex with its substrate2 ribulose-1,5-bisphosphate
|
| 74 | d1mzha_ |
|
not modelled |
12.0 |
6 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 75 | c2vwtA_ |
|
not modelled |
11.8 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:yfau, 2-keto-3-deoxy sugar aldolase;
PDBTitle: crystal structure of yfau, a metal ion dependent class ii2 aldolase from escherichia coli k12 - mg-pyruvate product3 complex
|
| 76 | d1vcva1 |
|
not modelled |
11.6 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 77 | c1bf2A_ |
|
not modelled |
11.3 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:isoamylase;
PDBTitle: structure of pseudomonas isoamylase
|
| 78 | d1vyra_ |
|
not modelled |
11.2 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 79 | c3c6cA_ |
|
not modelled |
11.1 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:3-keto-5-aminohexanoate cleavage enzyme;
PDBTitle: crystal structure of a putative 3-keto-5-aminohexanoate cleavage2 enzyme (reut_c6226) from ralstonia eutropha jmp134 at 1.72 a3 resolution
|
| 80 | d1jpma1 |
|
not modelled |
11.0 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
| 81 | d1m3ua_ |
|
not modelled |
10.8 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Ketopantoate hydroxymethyltransferase PanB |
| 82 | c2l69A_ |
|
not modelled |
10.7 |
12 |
PDB header:de novo protein
Chain: A: PDB Molecule:rossmann 2x3 fold protein;
PDBTitle: solution nmr structure of de novo designed protein, p-loop ntpase2 fold, northeast structural genomics consortium target or28
|
| 83 | c2gq8A_ |
|
not modelled |
10.6 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase, fmn-binding;
PDBTitle: structure of sye1, an oye homologue from s. ondeidensis, in complex2 with p-hydroxyacetophenone
|
| 84 | d2jnaa1 |
|
not modelled |
10.4 |
20 |
Fold:Dodecin subunit-like
Superfamily:YdgH-like
Family:YdgH-like |
| 85 | d1w3ia_ |
|
not modelled |
10.2 |
4 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 86 | d1gsaa1 |
|
not modelled |
10.1 |
12 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:Prokaryotic glutathione synthetase, N-terminal domain |
| 87 | d1q45a_ |
|
not modelled |
9.9 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 88 | c2uwqA_ |
|
not modelled |
9.8 |
5 |
PDB header:apoptosis
Chain: A: PDB Molecule:apoptosis-stimulating of p53 protein 2;
PDBTitle: solution structure of aspp2 n-terminus
|
| 89 | c3eypB_ |
|
not modelled |
9.8 |
12 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative alpha-l-fucosidase;
PDBTitle: crystal structure of putative alpha-l-fucosidase from bacteroides2 thetaiotaomicron
|
| 90 | d1m53a2 |
|
not modelled |
9.7 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 91 | d1vjia_ |
|
not modelled |
9.7 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 92 | c3l5aA_ |
|
not modelled |
9.7 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadh/flavin oxidoreductase/nadh oxidase;
PDBTitle: crystal structure of a probable nadh-dependent flavin oxidoreductase2 from staphylococcus aureus
|
| 93 | c3gkaB_ |
|
not modelled |
9.5 |
11 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:n-ethylmaleimide reductase;
PDBTitle: crystal structure of n-ethylmaleimidine reductase from2 burkholderia pseudomallei
|
| 94 | c1jd7A_ |
|
not modelled |
9.4 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-amylase;
PDBTitle: crystal structure analysis of the mutant k300r of2 pseudoalteromonas haloplanctis alpha-amylase
|
| 95 | d1vhxa_ |
|
not modelled |
9.4 |
15 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Putative Holliday junction resolvase RuvX |
| 96 | d1gwja_ |
|
not modelled |
9.3 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 97 | c3cqkB_ |
|
not modelled |
9.2 |
23 |
PDB header:isomerase
Chain: B: PDB Molecule:l-ribulose-5-phosphate 3-epimerase ulae;
PDBTitle: crystal structure of l-xylulose-5-phosphate 3-epimerase ulae (form b)2 complex with zn2+ and sulfate
|
| 98 | d1icpa_ |
|
not modelled |
9.1 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 99 | c3gr7A_ |
|
not modelled |
9.0 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadph dehydrogenase;
PDBTitle: structure of oye from geobacillus kaustophilus, hexagonal2 crystal form
|