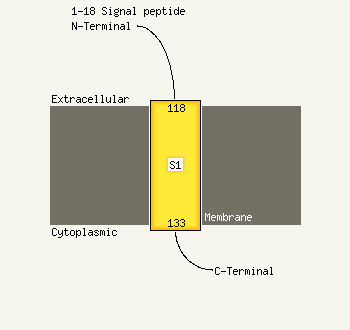
1 c2h1xB_
100.0
40
PDB header: hydrolaseChain: B: PDB Molecule: 5-hydroxyisourate hydrolase (formerly known asPDBTitle: crystal structure of 5-hydroxyisourate hydrolase (formerly2 known as trp, transthyretin related protein)
2 c2gpzC_
100.0
77
PDB header: hydrolaseChain: C: PDB Molecule: transthyretin-like protein;PDBTitle: transthyretin-like protein from salmonella dublin
3 d1ttaa_
100.0
32
Fold: Prealbumin-likeSuperfamily: Transthyretin (synonym: prealbumin)Family: Transthyretin (synonym: prealbumin)4 d1oo2a_
100.0
38
Fold: Prealbumin-likeSuperfamily: Transthyretin (synonym: prealbumin)Family: Transthyretin (synonym: prealbumin)5 d1kgia_
100.0
36
Fold: Prealbumin-likeSuperfamily: Transthyretin (synonym: prealbumin)Family: Transthyretin (synonym: prealbumin)6 d1f86a_
100.0
33
Fold: Prealbumin-likeSuperfamily: Transthyretin (synonym: prealbumin)Family: Transthyretin (synonym: prealbumin)7 d1tfpa_
100.0
39
Fold: Prealbumin-likeSuperfamily: Transthyretin (synonym: prealbumin)Family: Transthyretin (synonym: prealbumin)8 c2h0eA_
100.0
38
PDB header: hydrolaseChain: A: PDB Molecule: transthyretin-like protein pucm;PDBTitle: crystal structure of pucm in the absence of substrate
9 c3qvaB_
100.0
36
PDB header: hydrolaseChain: B: PDB Molecule: transthyretin-like protein;PDBTitle: structure of klebsiella pneumoniae 5-hydroxyisourate hydrolase
10 c2azqA_
94.5
25
PDB header: oxidoreductaseChain: A: PDB Molecule: catechol 1,2-dioxygenase;PDBTitle: crystal structure of catechol 1,2-dioxygenase from pseudomonas arvilla2 c-1
11 c3n9tA_
94.1
24
PDB header: oxidoreductaseChain: A: PDB Molecule: pnpc;PDBTitle: cryatal structure of hydroxyquinol 1,2-dioxygenase from pseudomonas2 putida dll-e4
12 c3kptA_
93.8
21
PDB header: cell adhesionChain: A: PDB Molecule: collagen adhesion protein;PDBTitle: crystal structure of bcpa, the major pilin subunit of2 bacillus cereus
13 d3pccm_
93.4
24
Fold: Prealbumin-likeSuperfamily: Aromatic compound dioxygenaseFamily: Aromatic compound dioxygenase14 c1tmxA_
92.7
13
PDB header: oxidoreductaseChain: A: PDB Molecule: hydroxyquinol 1,2-dioxygenase;PDBTitle: crystal structure of hydroxyquinol 1,2-dioxygenase from2 nocardioides simplex 3e
15 d1cwva1
92.3
21
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Invasin/intimin cell-adhesion fragmentsFamily: Invasin/intimin cell-adhesion fragments16 c2pz4A_
92.2
22
PDB header: cell adhesionChain: A: PDB Molecule: protein gbs052;PDBTitle: crystal structure of spab (gbs52), the minor pilin in gram-positive2 pathogen streptococcus agalactiae
17 d1h8la1
91.9
22
Fold: Prealbumin-likeSuperfamily: Carboxypeptidase regulatory domain-likeFamily: Carboxypeptidase regulatory domain18 c1h8lA_
91.8
22
PDB header: carboxypeptidaseChain: A: PDB Molecule: carboxypeptidase gp180 residues 503-882;PDBTitle: duck carboxypeptidase d domain ii in complex with gemsa
19 c3hj8A_
90.9
24
PDB header: oxidoreductaseChain: A: PDB Molecule: catechol 1,2-dioxygenase;PDBTitle: crystal structure determination of catechol 1,2-dioxygenase from2 rhodococcus opacus 1cp in complex with 4-chlorocatechol
20 d2burb1
90.7
22
Fold: Prealbumin-likeSuperfamily: Aromatic compound dioxygenaseFamily: Aromatic compound dioxygenase21 d1s9aa_
not modelled
90.6
15
Fold: Prealbumin-likeSuperfamily: Aromatic compound dioxygenaseFamily: Aromatic compound dioxygenase22 d1dmha_
not modelled
90.6
16
Fold: Prealbumin-likeSuperfamily: Aromatic compound dioxygenaseFamily: Aromatic compound dioxygenase23 c2boyC_
not modelled
90.4
28
PDB header: oxidoreductaseChain: C: PDB Molecule: 3-chlorocatechol 1,2-dioxygenase;PDBTitle: crystal structure of 3-chlorocatechol 1,2-dioxygenase from2 rhodococcus opacus 1cp
24 c3mn8A_
not modelled
89.4
27
PDB header: hydrolaseChain: A: PDB Molecule: lp15968p;PDBTitle: structure of drosophila melanogaster carboxypeptidase d isoform 1b2 short
25 c2xsuA_
not modelled
89.1
16
PDB header: oxidoreductaseChain: A: PDB Molecule: catechol 1,2 dioxygenase;PDBTitle: crystal structure of the a72g mutant of acinetobacter2 radioresistens catechol 1,2 dioxygenase
26 c3e8vA_
not modelled
88.9
30
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: possible transglutaminase-family protein;PDBTitle: crystal structure of a possible transglutaminase-family2 protein proteolytic fragment from bacteroides fragilis
27 c3irpX_
not modelled
88.0
33
PDB header: cell adhesionChain: X: PDB Molecule: uro-adherence factor a;PDBTitle: crystal structure of functional region of uafa from staphylococcus2 saprophyticus at 1.50 angstrom resolution
28 d1f00i1
not modelled
87.7
29
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Invasin/intimin cell-adhesion fragmentsFamily: Invasin/intimin cell-adhesion fragments29 d1nkga1
not modelled
85.5
19
Fold: Prealbumin-likeSuperfamily: Starch-binding domain-likeFamily: Rhamnogalacturonase B, RhgB, middle domain30 d1uwya1
not modelled
85.3
18
Fold: Prealbumin-likeSuperfamily: Carboxypeptidase regulatory domain-likeFamily: Carboxypeptidase regulatory domain31 c2nsmA_
not modelled
85.2
17
PDB header: hydrolaseChain: A: PDB Molecule: carboxypeptidase n catalytic chain;PDBTitle: crystal structure of the human carboxypeptidase n (kininase i)2 catalytic domain
32 c1uwyA_
not modelled
85.1
19
PDB header: hydrolaseChain: A: PDB Molecule: carboxypeptidase m;PDBTitle: crystal structure of human carboxypeptidase m
33 c2ww8A_
not modelled
79.9
13
PDB header: cell adhesionChain: A: PDB Molecule: cell wall surface anchor family protein;PDBTitle: structure of the pilus adhesin (rrga) from streptococcus2 pneumoniae
34 d2bura1
not modelled
79.8
19
Fold: Prealbumin-likeSuperfamily: Aromatic compound dioxygenaseFamily: Aromatic compound dioxygenase35 c2x5pA_
not modelled
78.3
19
PDB header: protein bindingChain: A: PDB Molecule: fibronectin binding protein;PDBTitle: crystal structure of the streptococcus pyogenes fibronectin binding2 protein fbab-b
36 c1cwvA_
not modelled
77.3
20
PDB header: structural proteinChain: A: PDB Molecule: invasin;PDBTitle: crystal structure of invasin: a bacterial integrin-binding protein
37 d3pcca_
not modelled
74.9
13
Fold: Prealbumin-likeSuperfamily: Aromatic compound dioxygenaseFamily: Aromatic compound dioxygenase38 c2p9rA_
not modelled
66.0
11
PDB header: signaling proteinChain: A: PDB Molecule: alpha-2-macroglobulin;PDBTitle: human alpha2-macroglogulin is composed of multiple domains,2 as predicted by homology with complement component c3
39 d1cwva2
not modelled
62.5
20
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Invasin/intimin cell-adhesion fragmentsFamily: Invasin/intimin cell-adhesion fragments40 c2xtlB_
not modelled
55.5
18
PDB header: structural proteinChain: B: PDB Molecule: cell wall surface anchor family protein;PDBTitle: structure of the major pilus backbone protein from streptococcus2 agalactiae
41 c3htlX_
not modelled
50.2
25
PDB header: structural protein, cell adhesionChain: X: PDB Molecule: putative surface-anchored fimbrial subunit;PDBTitle: structure of the corynebacterium diphtheriae major pilin2 spaa points to a modular pilus assembly with stabilizing3 isopeptide bonds
42 c2zxcA_
not modelled
48.2
16
PDB header: hydrolaseChain: A: PDB Molecule: neutral ceramidase;PDBTitle: seramidase complexed with c2
43 d1cwva3
not modelled
44.9
11
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Invasin/intimin cell-adhesion fragmentsFamily: Invasin/intimin cell-adhesion fragments44 d2dmca1
not modelled
42.7
8
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Filamin repeat (rod domain)45 d1ix2a_
not modelled
30.0
14
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Copper resistance protein C (CopC, PcoC)46 c2xicB_
not modelled
25.5
11
PDB header: cell adhesionChain: B: PDB Molecule: ancillary protein 1;PDBTitle: pilus-presented adhesin, spy0125 (cpa), p212121 form (esrf data)
47 d2yzca2
not modelled
21.7
25
Fold: T-foldSuperfamily: Tetrahydrobiopterin biosynthesis enzymes-likeFamily: Urate oxidase (uricase)48 c2x9yA_
not modelled
19.6
22
PDB header: cell adhesionChain: A: PDB Molecule: cell wall surface anchor family protein;PDBTitle: structure of the pilus backbone (rrgb) from streptococcus2 pneumoniae
49 c2yzbA_
not modelled
18.3
25
PDB header: oxidoreductaseChain: A: PDB Molecule: uricase;PDBTitle: crystal structure of uricase from arthrobacter globiformis2 in complex with uric acid (substrate)
50 c3hlkB_
not modelled
16.9
18
PDB header: hydrolaseChain: B: PDB Molecule: acyl-coenzyme a thioesterase 2, mitochondrial;PDBTitle: crystal structure of human mitochondrial acyl-coa2 thioesterase (acot2)
51 d2q3za1
not modelled
16.6
18
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Transglutaminase N-terminal domain52 c2v1lA_
not modelled
15.2
13
PDB header: unknown functionChain: A: PDB Molecule: hypothetical protein;PDBTitle: structure of the conserved hypothetical protein vc1805 from2 pathogenicity island vpi-2 of vibrio cholerae o1 biovar3 eltor str. n16961 shares structural homology with the4 human p32 protein
53 c2w56B_
not modelled
14.7
15
PDB header: unknown functionChain: B: PDB Molecule: vc0508;PDBTitle: structure of the hypothetical protein vc0508 from vibrio cholerae2 vsp-ii pathogenicity island
54 c3rpkA_
not modelled
12.9
22
PDB header: cell adhesionChain: A: PDB Molecule: backbone pilus subunit;PDBTitle: structure of the full-length major pilin rrgb from streptococcus2 pneumoniae
55 c2jmbA_
not modelled
12.7
26
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein atu4866;PDBTitle: solution structure of the protein atu4866 from agrobacterium2 tumefaciens
56 c3cu7A_
not modelled
12.4
17
PDB header: immune systemChain: A: PDB Molecule: complement c5;PDBTitle: human complement component 5
57 d2c9qa1
not modelled
12.2
18
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Copper resistance protein C (CopC, PcoC)58 d1vrma1
not modelled
12.0
53
Fold: T-foldSuperfamily: ApbE-likeFamily: ApbE-like59 c3lznA_
not modelled
11.7
17
PDB header: transport proteinChain: A: PDB Molecule: p19 protein;PDBTitle: crystal structure analysis of the apo p19 protein from campylobacter2 jejuni at 1.59 a at ph 9
60 d1ju3a1
not modelled
11.5
19
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: PepX C-terminal domain-like61 c2ougC_
not modelled
11.4
18
PDB header: transcriptionChain: C: PDB Molecule: transcriptional activator rfah;PDBTitle: crystal structure of the rfah transcription factor at 2.1a2 resolution
62 c2jh3C_
not modelled
11.0
26
PDB header: ribosomal proteinChain: C: PDB Molecule: ribosomal protein s2-related protein;PDBTitle: the crystal structure of dr2241 from deinococcus2 radiodurans at 1.9 a resolution reveals a multi-domain3 protein with structural similarity to chelatases but also4 with two additional novel domains
63 c2djcA_
not modelled
11.0
57
PDB header: cytokineChain: A: PDB Molecule: growth-blocking peptide;PDBTitle: solution structure of growth-blocking peptide of the2 tobacco cutworm, spodoptera litura
64 d1vjja1
not modelled
10.8
13
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Transglutaminase N-terminal domain65 d1olma2
not modelled
10.7
30
Fold: Supernatant protein factor (SPF), C-terminal domainSuperfamily: Supernatant protein factor (SPF), C-terminal domainFamily: Supernatant protein factor (SPF), C-terminal domain66 c1c8uA_
not modelled
10.5
14
PDB header: hydrolaseChain: A: PDB Molecule: acyl-coa thioesterase ii;PDBTitle: crystal structure of the e.coli thioesterase ii, a2 homologue of the human nef-binding enzyme
67 c1irrA_
not modelled
10.2
57
PDB header: cytokineChain: A: PDB Molecule: paralytic peptide;PDBTitle: solution structure of paralytic peptide of the silkworm,2 bombyx mori
68 c3pvmB_
not modelled
10.0
29
PDB header: immune systemChain: B: PDB Molecule: cobra venom factor;PDBTitle: structure of complement c5 in complex with cvf
69 c1hrlA_
not modelled
9.2
57
PDB header: toxinChain: A: PDB Molecule: paralytic peptide i;PDBTitle: structure of a paralytic peptide from an insect, manduca2 sexta
70 c1v28A_
not modelled
8.9
57
PDB header: toxinChain: A: PDB Molecule: paralytic peptide;PDBTitle: solution structure of paralytic peptide of the wild2 silkmoth, antheraea yamamai
71 d1qhda2
not modelled
8.6
16
Fold: Viral protein domainSuperfamily: Viral protein domainFamily: Top domain of virus capsid protein72 c2dj9A_
not modelled
8.6
57
PDB header: cytokineChain: A: PDB Molecule: growth-blocking peptide;PDBTitle: solution structure of growth-blocking peptide of the2 cabbage armyworm, mamestra brassicae
73 c2y7uM_
not modelled
8.3
18
PDB header: virusChain: M: PDB Molecule: coat protein;PDBTitle: x-ray structure of the grapevine fanleaf virus
74 d1zxqa1
not modelled
8.3
20
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: C2 set domains75 c2zxqA_
not modelled
8.1
24
PDB header: hydrolaseChain: A: PDB Molecule: endo-alpha-n-acetylgalactosaminidase;PDBTitle: crystal structure of endo-alpha-n-acetylgalactosaminidase2 from bifidobacterium longum (engbf)
76 c1nkgA_
not modelled
8.0
17
PDB header: lyaseChain: A: PDB Molecule: rhamnogalacturonase b;PDBTitle: rhamnogalacturonan lyase from aspergillus aculeatus
77 d1c8ua1
not modelled
7.8
14
Fold: Thioesterase/thiol ester dehydrase-isomeraseSuperfamily: Thioesterase/thiol ester dehydrase-isomeraseFamily: Acyl-CoA thioesterase78 c1o7dC_
not modelled
7.6
15
PDB header: hydrolaseChain: C: PDB Molecule: lysosomal alpha-mannosidase;PDBTitle: the structure of the bovine lysosomal a-mannosidase2 suggests a novel mechanism for low ph activation
79 d2g3ra1
not modelled
7.2
23
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: Tudor domain80 d1fcda2
not modelled
7.1
19
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains81 d1rm6a2
not modelled
7.1
20
Fold: Molybdenum cofactor-binding domainSuperfamily: Molybdenum cofactor-binding domainFamily: Molybdenum cofactor-binding domain82 d1z0mb1
not modelled
6.9
10
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: AMPK-beta glycogen binding domain-like83 d1t2sa_
not modelled
6.9
18
Fold: SH3-like barrelSuperfamily: PAZ domainFamily: PAZ domain84 c1zpuE_
not modelled
6.8
18
PDB header: oxidoreductaseChain: E: PDB Molecule: iron transport multicopper oxidase fet3;PDBTitle: crystal structure of fet3p, a multicopper oxidase that functions in2 iron import
85 d2vera1
not modelled
6.7
27
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Bacterial adhesinsFamily: Dr-family adhesin86 d1zdxa1
not modelled
6.4
21
Fold: FimD N-terminal domain-likeSuperfamily: FimD N-terminal domain-likeFamily: Usher N-domain87 c1fi8E_
not modelled
6.3
40
PDB header: hydrolase/hydrolase inhibitorChain: E: PDB Molecule: ecotin;PDBTitle: rat granzyme b [n66q] complexed to ecotin [81-84 iepd]
88 c1d2pA_
not modelled
6.3
24
PDB header: structural proteinChain: A: PDB Molecule: collagen adhesin;PDBTitle: crystal structure of two b repeat units (b1b2) of the2 collagen binding protein (cna) of staphylococcus aureus
89 d1v6ea_
not modelled
6.2
13
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: Ubiquitin-related90 c2o6dB_
not modelled
5.8
33
PDB header: membrane protein, protein bindingChain: B: PDB Molecule: 34 kda membrane antigen;PDBTitle: structure of native rtp34 from treponema pallidum
91 c1kv3F_
not modelled
5.7
17
PDB header: transferaseChain: F: PDB Molecule: protein-glutamine gamma-glutamyltransferase;PDBTitle: human tissue transglutaminase in gdp bound form
92 d1eala_
not modelled
5.5
15
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like93 c1t3qB_
not modelled
5.5
34
PDB header: oxidoreductaseChain: B: PDB Molecule: quinoline 2-oxidoreductase large subunit;PDBTitle: crystal structure of quinoline 2-oxidoreductase from pseudomonas2 putida 86
94 d1m7xa2
not modelled
5.4
15
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain95 c3nrqB_
not modelled
5.4
25
PDB header: transport proteinChain: B: PDB Molecule: periplasmic protein-probably involved in high-affinity fe2+PDBTitle: crystal structure of copper-reconstituted fetp from uropathogenic2 escherichia coli strain f11
96 d1a9xa1
not modelled
5.4
63
Fold: Carbamoyl phosphate synthetase, large subunit connection domainSuperfamily: Carbamoyl phosphate synthetase, large subunit connection domainFamily: Carbamoyl phosphate synthetase, large subunit connection domain97 c3ecqA_
not modelled
5.3
19
PDB header: hydrolaseChain: A: PDB Molecule: endo-alpha-n-acetylgalactosaminidase;PDBTitle: endo-alpha-n-acetylgalactosaminidase from streptococcus pneumoniae:2 semet structure
98 d1lnsa2
not modelled
5.3
9
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: PepX C-terminal domain-like99 d2p0ma2
not modelled
5.3
40
Fold: Lipase/lipooxygenase domain (PLAT/LH2 domain)Superfamily: Lipase/lipooxygenase domain (PLAT/LH2 domain)Family: Lipoxigenase N-terminal domain