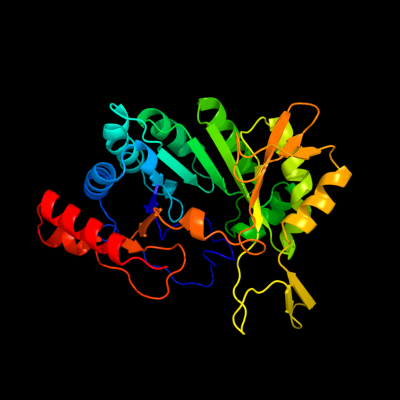
| 1 | d1tv8a_
|
|
 |
100.0 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Radical SAM enzymes
Family:MoCo biosynthesis proteins |
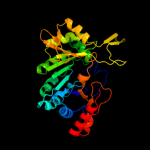
| 2 | c2yx0A_
|
|
 |
99.9 |
13 |
PDB header:metal binding protein
Chain: A: PDB Molecule:radical sam enzyme;
PDBTitle: crystal structure of p. horikoshii tyw1
|
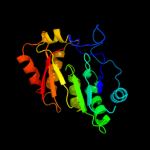
| 3 | c3c8fA_
|
|
 |
99.8 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pyruvate formate-lyase 1-activating enzyme;
PDBTitle: 4fe-4s-pyruvate formate-lyase activating enzyme with2 partially disordered adomet
|
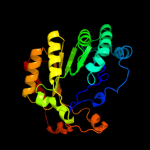
| 4 | c2a5hC_
|
|
 |
99.6 |
15 |
PDB header:isomerase
Chain: C: PDB Molecule:l-lysine 2,3-aminomutase;
PDBTitle: 2.1 angstrom x-ray crystal structure of lysine-2,3-aminomutase from2 clostridium subterminale sb4, with michaelis analog (l-alpha-lysine3 external aldimine form of pyridoxal-5'-phosphate).
|
| 5 | d1olta_
|
|
 |
99.6 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:Radical SAM enzymes
Family:Oxygen-independent coproporphyrinogen III oxidase HemN |
| 6 | c1r30A_
|
|
 |
99.6 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:biotin synthase;
PDBTitle: the crystal structure of biotin synthase, an s-2 adenosylmethionine-dependent radical enzyme
|
| 7 | d1r30a_
|
|
 |
99.6 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Radical SAM enzymes
Family:Biotin synthase |
| 8 | c3t7vA_
|
|
 |
99.5 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:methylornithine synthase pylb;
PDBTitle: crystal structure of methylornithine synthase (pylb)
|
| 9 | c3cixA_
|
|
 |
99.5 |
12 |
PDB header:adomet binding protein
Chain: A: PDB Molecule:fefe-hydrogenase maturase;
PDBTitle: x-ray structure of the [fefe]-hydrogenase maturase hyde from2 thermotoga maritima in complex with thiocyanate
|
| 10 | c2z2uA_
|
|
 |
99.5 |
20 |
PDB header:metal binding protein
Chain: A: PDB Molecule:upf0026 protein mj0257;
PDBTitle: crystal structure of archaeal tyw1
|
| 11 | c3canA_
|
|
 |
99.5 |
15 |
PDB header:lyase activator
Chain: A: PDB Molecule:pyruvate-formate lyase-activating enzyme;
PDBTitle: crystal structure of a domain of pyruvate-formate lyase-activating2 enzyme from bacteroides vulgatus atcc 8482
|
| 12 | c3rfaA_
|
|
 |
99.4 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ribosomal rna large subunit methyltransferase n;
PDBTitle: x-ray structure of rlmn from escherichia coli in complex with s-2 adenosylmethionine
|
| 13 | c2qgqF_
|
|
 |
98.9 |
9 |
PDB header:structural genomics, unknown function
Chain: F: PDB Molecule:protein tm_1862;
PDBTitle: crystal structure of tm_1862 from thermotoga maritima.2 northeast structural genomics consortium target vr77
|
| 14 | c3ivuB_
|
|
 |
97.2 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:homocitrate synthase, mitochondrial;
PDBTitle: homocitrate synthase lys4 bound to 2-og
|
| 15 | c3bleA_
|
|
 |
96.9 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:citramalate synthase from leptospira interrogans;
PDBTitle: crystal structure of the catalytic domain of licms in2 complexed with malonate
|
| 16 | c3ewbX_
|
|
 |
96.9 |
11 |
PDB header:transferase
Chain: X: PDB Molecule:2-isopropylmalate synthase;
PDBTitle: crystal structure of n-terminal domain of putative 2-2 isopropylmalate synthase from listeria monocytogenes
|
| 17 | c2ftpA_
|
|
 |
96.8 |
10 |
PDB header:lyase
Chain: A: PDB Molecule:hydroxymethylglutaryl-coa lyase;
PDBTitle: crystal structure of hydroxymethylglutaryl-coa lyase from pseudomonas2 aeruginosa
|
| 18 | c2cw6B_
|
|
 |
95.9 |
11 |
PDB header:lyase
Chain: B: PDB Molecule:hydroxymethylglutaryl-coa lyase, mitochondrial;
PDBTitle: crystal structure of human hmg-coa lyase: insights into2 catalysis and the molecular basis for3 hydroxymethylglutaric aciduria
|
| 19 | c1ydoC_
|
|
 |
95.8 |
12 |
PDB header:lyase
Chain: C: PDB Molecule:hmg-coa lyase;
PDBTitle: crystal structure of the bacillis subtilis hmg-coa lyase, northeast2 structural genomics target sr181.
|
| 20 | c1ydnA_
|
|
 |
95.4 |
7 |
PDB header:lyase
Chain: A: PDB Molecule:hydroxymethylglutaryl-coa lyase;
PDBTitle: crystal structure of the hmg-coa lyase from brucella melitensis,2 northeast structural genomics target lr35.
|
| 21 | c3eegB_ |
|
not modelled |
93.7 |
8 |
PDB header:transferase
Chain: B: PDB Molecule:2-isopropylmalate synthase;
PDBTitle: crystal structure of a 2-isopropylmalate synthase from2 cytophaga hutchinsonii
|
| 22 | c1nvmG_ |
|
not modelled |
85.9 |
8 |
PDB header:lyase/oxidoreductase
Chain: G: PDB Molecule:4-hydroxy-2-oxovalerate aldolase;
PDBTitle: crystal structure of a bifunctional aldolase-dehydrogenase :2 sequestering a reactive and volatile intermediate
|
| 23 | c1sr9A_ |
|
not modelled |
84.6 |
9 |
PDB header:transferase
Chain: A: PDB Molecule:2-isopropylmalate synthase;
PDBTitle: crystal structure of leua from mycobacterium tuberculosis
|
| 24 | c2qv5A_ |
|
not modelled |
72.7 |
9 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein atu2773;
PDBTitle: crystal structure of uncharacterized protein atu2773 from2 agrobacterium tumefaciens c58
|
| 25 | d1nvma2 |
|
not modelled |
70.9 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:HMGL-like |
| 26 | c1rr2A_ |
|
not modelled |
70.7 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:transcarboxylase 5s subunit;
PDBTitle: propionibacterium shermanii transcarboxylase 5s subunit bound to 2-2 ketobutyric acid
|
| 27 | c2zyfA_ |
|
not modelled |
69.1 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:homocitrate synthase;
PDBTitle: crystal structure of homocitrate synthase from thermus thermophilus2 complexed with magnesuim ion and alpha-ketoglutarate
|
| 28 | c3dxiB_ |
|
not modelled |
65.9 |
9 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative aldolase;
PDBTitle: crystal structure of the n-terminal domain of a putative2 aldolase (bvu_2661) from bacteroides vulgatus
|
| 29 | c2vefB_ |
|
not modelled |
59.6 |
9 |
PDB header:transferase
Chain: B: PDB Molecule:dihydropteroate synthase;
PDBTitle: dihydropteroate synthase from streptococcus pneumoniae
|
| 30 | c2nx9B_ |
|
not modelled |
57.2 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:oxaloacetate decarboxylase 2, subunit alpha;
PDBTitle: crystal structure of the carboxyltransferase domain of the2 oxaloacetate decarboxylase na+ pump from vibrio cholerae
|
| 31 | d2nlya1 |
|
not modelled |
55.1 |
9 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycoside hydrolase/deacetylase
Family:Divergent polysaccharide deacetylase |
| 32 | c3iwpK_ |
|
not modelled |
54.9 |
11 |
PDB header:metal binding protein
Chain: K: PDB Molecule:copper homeostasis protein cutc homolog;
PDBTitle: crystal structure of human copper homeostasis protein cutc
|
| 33 | c2zq0B_ |
|
not modelled |
54.3 |
11 |
PDB header:hydrolase
Chain: B: PDB Molecule:alpha-glucosidase (alpha-glucosidase susb);
PDBTitle: crystal structure of susb complexed with acarbose
|
| 34 | c2bdqA_ |
|
not modelled |
54.2 |
11 |
PDB header:metal transport
Chain: A: PDB Molecule:copper homeostasis protein cutc;
PDBTitle: crystal structure of the putative copper homeostasis2 protein cutc from streptococcus agalactiae, northeast3 strucural genomics target sar15.
|
| 35 | d1muwa_ |
|
not modelled |
53.7 |
8 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 36 | c3hpxB_ |
|
not modelled |
52.5 |
9 |
PDB header:transferase
Chain: B: PDB Molecule:2-isopropylmalate synthase;
PDBTitle: crystal structure of mycobacterium tuberculosis leua active site2 domain 1-425 (truncation mutant delta:426-644)
|
| 37 | d1ajza_ |
|
not modelled |
51.5 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 38 | c3czkA_ |
|
not modelled |
50.3 |
8 |
PDB header:hydrolase
Chain: A: PDB Molecule:sucrose hydrolase;
PDBTitle: crystal structure analysis of sucrose hydrolase(suh) e322q-2 sucrose complex
|
| 39 | d1tx2a_ |
|
not modelled |
50.1 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 40 | c1tx2A_ |
|
not modelled |
50.1 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:dhps, dihydropteroate synthase;
PDBTitle: dihydropteroate synthetase, with bound inhibitor manic, from bacillus2 anthracis
|
| 41 | d1g5aa2 |
|
not modelled |
49.8 |
8 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 42 | c3bg3B_ |
|
not modelled |
47.8 |
14 |
PDB header:ligase
Chain: B: PDB Molecule:pyruvate carboxylase, mitochondrial;
PDBTitle: crystal structure of human pyruvate carboxylase (missing2 the biotin carboxylase domain at the n-terminus)
|
| 43 | d2zdra2 |
|
not modelled |
47.6 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:NeuB-like |
| 44 | c1jgiA_ |
|
not modelled |
47.5 |
8 |
PDB header:transferase
Chain: A: PDB Molecule:amylosucrase;
PDBTitle: crystal structure of the active site mutant glu328gln of2 amylosucrase from neisseria polysaccharea in complex with3 the natural substrate sucrose
|
| 45 | c3n2xB_ |
|
not modelled |
46.8 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:uncharacterized protein yage;
PDBTitle: crystal structure of yage, a prophage protein belonging to the2 dihydrodipicolinic acid synthase family from e. coli k12 in complex3 with pyruvate
|
| 46 | c3ol0C_ |
|
not modelled |
45.0 |
14 |
PDB header:de novo protein
Chain: C: PDB Molecule:de novo designed monomer trefoil-fold sub-domain which
PDBTitle: crystal structure of monofoil-4p homo-trimer: de novo designed monomer2 trefoil-fold sub-domain which forms homo-trimer assembly
|
| 47 | c2knpA_ |
|
not modelled |
44.9 |
43 |
PDB header:unknown function
Chain: A: PDB Molecule:mcocc-1;
PDBTitle: isolation and characterization of peptides from momordica2 cochinchinensis seeds.
|
| 48 | d1twda_ |
|
not modelled |
44.3 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:CutC-like
Family:CutC-like |
| 49 | d1yhta1 |
|
not modelled |
40.9 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-N-acetylhexosaminidase catalytic domain |
| 50 | c3dz1A_ |
|
not modelled |
39.1 |
11 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 1.87a resolution
|
| 51 | c3chvA_ |
|
not modelled |
36.8 |
13 |
PDB header:metal binding protein
Chain: A: PDB Molecule:prokaryotic domain of unknown function (duf849) with a tim
PDBTitle: crystal structure of a prokaryotic domain of unknown function (duf849)2 member (spoa0042) from silicibacter pomeroyi dss-3 at 1.45 a3 resolution
|
| 52 | c2gjxE_ |
|
not modelled |
35.3 |
9 |
PDB header:hydrolase
Chain: E: PDB Molecule:beta-hexosaminidase alpha chain;
PDBTitle: crystallographic structure of human beta-hexosaminidase a
|
| 53 | c2ekcA_ |
|
not modelled |
33.8 |
11 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: structural study of project id aq_1548 from aquifex aeolicus vf5
|
| 54 | c3hf3A_ |
|
not modelled |
33.7 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:chromate reductase;
PDBTitle: old yellow enzyme from thermus scotoductus sa-01
|
| 55 | c3lciA_ |
|
not modelled |
31.1 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:n-acetylneuraminate lyase;
PDBTitle: the d-sialic acid aldolase mutant v251w
|
| 56 | d1qwga_ |
|
not modelled |
30.5 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:(2r)-phospho-3-sulfolactate synthase ComA
Family:(2r)-phospho-3-sulfolactate synthase ComA |
| 57 | c3cprB_ |
|
not modelled |
29.9 |
11 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthetase;
PDBTitle: the crystal structure of corynebacterium glutamicum2 dihydrodipicolinate synthase to 2.2 a resolution
|
| 58 | d1qt1a_ |
|
not modelled |
29.8 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 59 | c2v9dB_ |
|
not modelled |
26.9 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:yage;
PDBTitle: crystal structure of yage, a prophage protein belonging to2 the dihydrodipicolinic acid synthase family from e. coli3 k12
|
| 60 | c3navB_ |
|
not modelled |
26.5 |
11 |
PDB header:lyase
Chain: B: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of an alpha subunit of tryptophan synthase from2 vibrio cholerae o1 biovar el tor str. n16961
|
| 61 | d2a6na1 |
|
not modelled |
26.2 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 62 | d1v93a_ |
|
not modelled |
25.5 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:FAD-linked oxidoreductase
Family:Methylenetetrahydrofolate reductase |
| 63 | c1ps9A_ |
|
not modelled |
25.3 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2,4-dienoyl-coa reductase;
PDBTitle: the crystal structure and reaction mechanism of e. coli 2,4-2 dienoyl coa reductase
|
| 64 | c3rpmA_ |
|
not modelled |
25.2 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-n-acetyl-hexosaminidase;
PDBTitle: crystal structure of the first gh20 domain of a novel beta-n-acetyl-2 hexosaminidase strh from streptococcus pneumoniae r6
|
| 65 | c3e02A_ |
|
not modelled |
25.2 |
11 |
PDB header:metal binding protein
Chain: A: PDB Molecule:uncharacterized protein duf849;
PDBTitle: crystal structure of a duf849 family protein (bxe_c0271) from2 burkholderia xenovorans lb400 at 1.90 a resolution
|
| 66 | d1hl2a_ |
|
not modelled |
25.1 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 67 | d1ad1a_ |
|
not modelled |
25.1 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 68 | c1nouA_ |
|
not modelled |
25.0 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-hexosaminidase beta chain;
PDBTitle: native human lysosomal beta-hexosaminidase isoform b
|
| 69 | d1m53a2 |
|
not modelled |
24.3 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 70 | c1qysA_ |
|
not modelled |
24.2 |
13 |
PDB header:de novo protein
Chain: A: PDB Molecule:top7;
PDBTitle: crystal structure of top7: a computationally designed2 protein with a novel fold
|
| 71 | c2ylaA_ |
|
not modelled |
24.1 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-n-acetylhexosaminidase;
PDBTitle: inhibition of the pneumococcal virulence factor strh and2 molecular insights into n-glycan recognition and3 hydrolysis
|
| 72 | c2g7zB_ |
|
not modelled |
23.8 |
5 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:conserved hypothetical protein spy1493;
PDBTitle: conserved degv-like protein of unknown function from streptococcus2 pyogenes m1 gas binds long-chain fatty acids
|
| 73 | c3khdC_ |
|
not modelled |
23.6 |
7 |
PDB header:transferase
Chain: C: PDB Molecule:pyruvate kinase;
PDBTitle: crystal structure of pff1300w.
|
| 74 | d1nowa1 |
|
not modelled |
22.1 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-N-acetylhexosaminidase catalytic domain |
| 75 | d1z41a1 |
|
not modelled |
22.1 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 76 | c2ou4C_ |
|
not modelled |
21.1 |
9 |
PDB header:isomerase
Chain: C: PDB Molecule:d-tagatose 3-epimerase;
PDBTitle: crystal structure of d-tagatose 3-epimerase from2 pseudomonas cichorii
|
| 77 | c3b4uB_ |
|
not modelled |
20.6 |
11 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from agrobacterium2 tumefaciens str. c58
|
| 78 | c2qmaB_ |
|
not modelled |
20.4 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:diaminobutyrate-pyruvate transaminase and l-2,4-
PDBTitle: crystal structure of glutamate decarboxylase domain of2 diaminobutyrate-pyruvate transaminase and l-2,4-diaminobutyrate3 decarboxylase from vibrio parahaemolyticus
|
| 79 | c3g0sA_ |
|
not modelled |
20.1 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: dihydrodipicolinate synthase from salmonella typhimurium lt2
|
| 80 | c3fluD_ |
|
not modelled |
20.0 |
10 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from the pathogen2 neisseria meningitidis
|
| 81 | d1uwva2 |
|
not modelled |
19.9 |
45 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:(Uracil-5-)-methyltransferase |
| 82 | d1eyea_ |
|
not modelled |
19.8 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 83 | c3pueA_ |
|
not modelled |
19.7 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of the complex of dhydrodipicolinate synthase from2 acinetobacter baumannii with lysine at 2.6a resolution
|
| 84 | c2jvfA_ |
|
not modelled |
19.6 |
17 |
PDB header:de novo protein
Chain: A: PDB Molecule:de novo protein m7;
PDBTitle: solution structure of m7, a computationally-designed2 artificial protein
|
| 85 | c3qfeB_ |
|
not modelled |
19.6 |
8 |
PDB header:lyase
Chain: B: PDB Molecule:putative dihydrodipicolinate synthase family protein;
PDBTitle: crystal structures of a putative dihydrodipicolinate synthase family2 protein from coccidioides immitis
|
| 86 | c3lmyA_ |
|
not modelled |
19.3 |
8 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-hexosaminidase subunit beta;
PDBTitle: the crystal structure of beta-hexosaminidase b in complex with2 pyrimethamine
|
| 87 | c1xuzA_ |
|
not modelled |
19.0 |
16 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:polysialic acid capsule biosynthesis protein siac;
PDBTitle: crystal structure analysis of sialic acid synthase (neub)from2 neisseria meningitidis, bound to mn2+, phosphoenolpyruvate, and n-3 acetyl mannosaminitol
|
| 88 | c2dh3A_ |
|
not modelled |
18.6 |
21 |
PDB header:transport protein, signaling protein
Chain: A: PDB Molecule:4f2 cell-surface antigen heavy chain;
PDBTitle: crystal structure of human ed-4f2hc
|
| 89 | c3ogfA_ |
|
not modelled |
18.3 |
10 |
PDB header:de novo protein
Chain: A: PDB Molecule:de novo designed dimeric trefoil-fold sub-domain which
PDBTitle: crystal structure of difoil-4p homo-trimer: de novo designed dimeric2 trefoil-fold sub-domain which forms homo-trimer assembly
|
| 90 | c3ct7E_ |
|
not modelled |
17.5 |
11 |
PDB header:isomerase
Chain: E: PDB Molecule:d-allulose-6-phosphate 3-epimerase;
PDBTitle: crystal structure of d-allulose 6-phosphate 3-epimerase2 from escherichia coli k-12
|
| 91 | c3lppA_ |
|
not modelled |
16.7 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:sucrase-isomaltase;
PDBTitle: crystal complex of n-terminal sucrase-isomaltase with kotalanol
|
| 92 | c3rcnA_ |
|
not modelled |
16.7 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-n-acetylhexosaminidase;
PDBTitle: crystal structure of beta-n-acetylhexosaminidase from arthrobacter2 aurescens
|
| 93 | c3no5C_ |
|
not modelled |
16.4 |
15 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a pfam duf849 domain containing protein2 (reut_a1631) from ralstonia eutropha jmp134 at 1.90 a resolution
|
| 94 | c1uwvA_ |
|
not modelled |
16.0 |
50 |
PDB header:transferase
Chain: A: PDB Molecule:23s rrna (uracil-5-)-methyltransferase ruma;
PDBTitle: crystal structure of ruma, the iron-sulfur cluster2 containing e. coli 23s ribosomal rna 5-methyluridine3 methyltransferase
|
| 95 | d1qbaa3 |
|
not modelled |
15.6 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-N-acetylhexosaminidase catalytic domain |
| 96 | c2ehhE_ |
|
not modelled |
15.6 |
16 |
PDB header:lyase
Chain: E: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 aquifex aeolicus
|
| 97 | d1o5ka_ |
|
not modelled |
15.5 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 98 | c3daqB_ |
|
not modelled |
15.4 |
9 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from methicillin-2 resistant staphylococcus aureus
|
| 99 | c2rfgB_ |
|
not modelled |
15.3 |
9 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from hahella2 chejuensis at 1.5a resolution
|