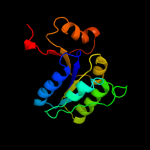
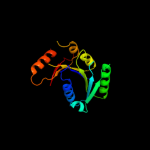
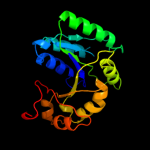
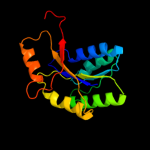
1 c1nijA_
100.0
25
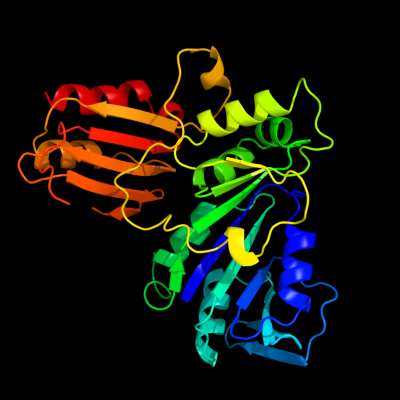
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein yjia;PDBTitle: yjia protein
2 d1nija1
100.0
29
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like3 c2hf9A_
99.7
18
PDB header: hydrolase, metal binding proteinChain: A: PDB Molecule: probable hydrogenase nickel incorporationPDBTitle: crystal structure of hypb from methanocaldococcus2 jannaschii in the triphosphate form
4 d1nija2
99.7
16
Fold: Hypothetical protein YjiA, C-terminal domainSuperfamily: Hypothetical protein YjiA, C-terminal domainFamily: Hypothetical protein YjiA, C-terminal domain5 c2wsmB_
99.5
19
PDB header: metal binding proteinChain: B: PDB Molecule: hydrogenase expression/formation protein (hypb);PDBTitle: crystal structure of hydrogenase maturation factor hypb from2 archaeoglobus fulgidus
6 c3nxsA_
99.5
21
PDB header: transport proteinChain: A: PDB Molecule: lao/ao transport system atpase;PDBTitle: crystal structure of lao/ao transport system from mycobacterium2 smegmatis bound to gdp
7 d1yrba1
99.5
20
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like8 d2qm8a1
99.3
26
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like9 c2f1rA_
99.3
15
PDB header: biosynthetic proteinChain: A: PDB Molecule: molybdopterin-guanine dinucleotide biosynthesisPDBTitle: crystal structure of molybdopterin-guanine biosynthesis2 protein b (mobb)
10 d1qzxa3
99.2
14
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like11 c3md0A_
99.1
22
PDB header: transport proteinChain: A: PDB Molecule: arginine/ornithine transport system atpase;PDBTitle: crystal structure of arginine/ornithine transport system2 atpase from mycobacterium tuberculosis bound to gdp (a ras-3 like gtpase superfamily protein)
12 c3dmdA_
99.1
19
PDB header: transport proteinChain: A: PDB Molecule: signal recognition particle receptor;PDBTitle: structures and conformations in solution of the signal recognition2 particle receptor from the archaeon pyrococcus furiosus
13 d2p67a1
99.1
19
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like14 d1ls1a2
99.0
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like15 c2cnwF_
99.0
16
PDB header: signal recognitionChain: F: PDB Molecule: cell division protein ftsy;PDBTitle: gdpalf4 complex of the srp gtpases ffh and ftsy
16 c3dm5A_
99.0
16
PDB header: rna binding protein, transport proteinChain: A: PDB Molecule: signal recognition 54 kda protein;PDBTitle: structures of srp54 and srp19, the two proteins assembling2 the ribonucleic core of the signal recognition particle3 from the archaeon pyrococcus furiosus.
17 c1zu4A_
98.9
16
PDB header: protein transportChain: A: PDB Molecule: ftsy;PDBTitle: crystal structure of ftsy from mycoplasma mycoides- space2 group p21212
18 c2j7pA_
98.9
18
PDB header: signal recognitionChain: A: PDB Molecule: signal recognition particle protein;PDBTitle: gmppnp-stabilized ng domain complex of the srp gtpases ffh2 and ftsy
19 c2iy3A_
98.8
19
PDB header: rna-bindingChain: A: PDB Molecule: signal recognition particle protein ffh;PDBTitle: structure of the e. coli signal recognition particle2 bound to a translating ribosome
20 c2wwwB_
98.8
20
PDB header: transport proteinChain: B: PDB Molecule: methylmalonic aciduria type a protein,PDBTitle: crystal structure of methylmalonic acidemia type a protein
21 d1vmaa2
not modelled
98.8
15
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like22 c2yhsA_
not modelled
98.8
15
PDB header: cell cycleChain: A: PDB Molecule: cell division protein ftsy;PDBTitle: structure of the e. coli srp receptor ftsy
23 c2v3cC_
not modelled
98.8
17
PDB header: signaling proteinChain: C: PDB Molecule: signal recognition 54 kda protein;PDBTitle: crystal structure of the srp54-srp19-7s.s srp rna complex2 of m. jannaschii
24 c2qy9A_
not modelled
98.8
15
PDB header: protein transportChain: A: PDB Molecule: cell division protein ftsy;PDBTitle: structure of the ng+1 construct of the e. coli srp receptor2 ftsy
25 c3b9qA_
not modelled
98.7
16
PDB header: protein transportChain: A: PDB Molecule: chloroplast srp receptor homolog, alpha subunitPDBTitle: the crystal structure of cpftsy from arabidopsis thaliana
26 c2og2A_
not modelled
98.7
17
PDB header: protein transportChain: A: PDB Molecule: putative signal recognition particle receptor;PDBTitle: crystal structure of chloroplast ftsy from arabidopsis2 thaliana
27 c2j37W_
not modelled
98.7
11
PDB header: ribosomeChain: W: PDB Molecule: signal recognition particle 54 kda proteinPDBTitle: model of mammalian srp bound to 80s rncs
28 d2qy9a2
not modelled
98.7
13
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like29 c1vmaA_
not modelled
98.7
16
PDB header: protein transportChain: A: PDB Molecule: cell division protein ftsy;PDBTitle: crystal structure of cell division protein ftsy (tm0570) from2 thermotoga maritima at 1.60 a resolution
30 d1okkd2
not modelled
98.5
18
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like31 c2e87A_
not modelled
98.5
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein ph1320;PDBTitle: crystal structure of hypothetical gtp-binding protein ph1320 from2 pyrococcus horikoshii ot3, in complex with gdp
32 c2qptA_
not modelled
98.5
19
PDB header: endocytosisChain: A: PDB Molecule: eh domain-containing protein-2;PDBTitle: crystal structure of an ehd atpase involved in membrane remodelling
33 d1xjca_
not modelled
98.4
20
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like34 c2px0D_
not modelled
98.4
15
PDB header: biosynthetic proteinChain: D: PDB Molecule: flagellar biosynthesis protein flhf;PDBTitle: crystal structure of flhf complexed with gmppnp/mg(2+)
35 c2gzaB_
not modelled
98.3
27
PDB header: hydrolaseChain: B: PDB Molecule: type iv secretion system protein virb11;PDBTitle: crystal structure of the virb11 atpase from the brucella suis type iv2 secretion system in complex with sulphate
36 c1qzwC_
not modelled
98.3
16
PDB header: signaling protein/rnaChain: C: PDB Molecule: signal recognition 54 kda protein;PDBTitle: crystal structure of the complete core of archaeal srp and2 implications for inter-domain communication
37 c1egaB_
not modelled
98.3
13
PDB header: hydrolaseChain: B: PDB Molecule: protein (gtp-binding protein era);PDBTitle: crystal structure of a widely conserved gtpase era
38 c2qthA_
not modelled
98.3
16
PDB header: nucleotide binding proteinChain: A: PDB Molecule: gtp-binding protein;PDBTitle: crystal structure of a gtp-binding protein from the2 hyperthermophilic archaeon sulfolobus solfataricus in3 complex with gdp
39 d1g6oa_
not modelled
98.3
20
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)40 c1wb1C_
not modelled
98.2
19
PDB header: protein synthesisChain: C: PDB Molecule: translation elongation factor selb;PDBTitle: crystal structure of translation elongation factor selb2 from methanococcus maripaludis in complex with gdp
41 c2j289_
not modelled
98.2
19
PDB header: ribosomeChain: 9: PDB Molecule: signal recognition particle 54;PDBTitle: model of e. coli srp bound to 70s rncs
42 c2qa5A_
not modelled
98.2
17
PDB header: cell cycle, structural proteinChain: A: PDB Molecule: septin-2;PDBTitle: crystal structure of sept2 g-domain
43 c2gedB_
not modelled
98.1
18
PDB header: protein transport, signaling proteinChain: B: PDB Molecule: signal recognition particle receptor betaPDBTitle: signal recognition particle receptor beta-subunit in2 nucleotide-free dimerized form
44 c2oaq1_
not modelled
98.1
22
PDB header: hydrolaseChain: 1: PDB Molecule: type ii secretion system protein;PDBTitle: crystal structure of the archaeal secretion atpase gspe in complex2 with phosphate
45 c1kk3A_
not modelled
98.1
16
PDB header: translationChain: A: PDB Molecule: eif2gamma;PDBTitle: structure of the wild-type large gamma subunit of2 initiation factor eif2 from pyrococcus abyssi complexed3 with gdp-mg2+
46 d1np6a_
not modelled
98.0
18
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like47 c1d2eA_
not modelled
98.0
11
PDB header: rna binding proteinChain: A: PDB Molecule: elongation factor tu (ef-tu);PDBTitle: crystal structure of mitochondrial ef-tu in complex with gdp
48 c3t5dC_
not modelled
98.0
19
PDB header: signaling proteinChain: C: PDB Molecule: septin-7;PDBTitle: crystal structure of septin 7 in complex with gdp
49 c2plfA_
not modelled
97.9
16
PDB header: translationChain: A: PDB Molecule: translation initiation factor 2 gamma subunit;PDBTitle: the structure of aif2gamma subunit from the archaeon2 sulfolobus solfataricus in the nucleotide-free form.
50 c3jvvA_
not modelled
97.9
23
PDB header: atp binding proteinChain: A: PDB Molecule: twitching mobility protein;PDBTitle: crystal structure of p. aeruginosa pilt with bound amp-pcp
51 c2qagB_
not modelled
97.9
23
PDB header: cell cycle, structural proteinChain: B: PDB Molecule: septin-6;PDBTitle: crystal structure of human septin trimer 2/6/7
52 c1wf3A_
not modelled
97.9
14
PDB header: hydrolaseChain: A: PDB Molecule: gtp-binding protein;PDBTitle: crystal structure of gtp-binding protein tt1341 from thermus2 thermophilus hb8
53 d1p9ra_
not modelled
97.9
28
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)54 c3gehA_
not modelled
97.8
17
PDB header: hydrolaseChain: A: PDB Molecule: trna modification gtpase mnme;PDBTitle: crystal structure of mnme from nostoc in complex with gdp, folinic2 acid and zn
55 c2bvnB_
not modelled
97.8
11
PDB header: elongation factorChain: B: PDB Molecule: elongation factor tu;PDBTitle: e. coli ef-tu:gdpnp in complex with the antibiotic2 enacyloxin iia
56 c3k53B_
not modelled
97.8
19
PDB header: metal transportChain: B: PDB Molecule: ferrous iron transport protein b;PDBTitle: crystal structure of nfeob from p. furiosus
57 c3ftqA_
not modelled
97.8
19
PDB header: cell cycleChain: A: PDB Molecule: septin-2;PDBTitle: crystal structure of septin 2 in complex with gppnhp and2 mg2+
58 d2c78a3
not modelled
97.8
11
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins59 c1mkyA_
not modelled
97.8
20
PDB header: ligand binding proteinChain: A: PDB Molecule: probable gtp-binding protein enga;PDBTitle: structural analysis of the domain interactions in der, a2 switch protein containing two gtpase domains
60 d2dy1a2
not modelled
97.8
20
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins61 c2qagC_
not modelled
97.8
23
PDB header: cell cycle, structural proteinChain: C: PDB Molecule: septin-7;PDBTitle: crystal structure of human septin trimer 2/6/7
62 c2xtpA_
not modelled
97.8
13
PDB header: immune systemChain: A: PDB Molecule: gtpase imap family member 2;PDBTitle: crystal structure of nucleotide-free human gimap2, amino2 acid residues 1-260
63 c1xzqA_
not modelled
97.8
23
PDB header: hydrolaseChain: A: PDB Molecule: probable trna modification gtpase trme;PDBTitle: structure of the gtp-binding protein trme from thermotoga2 maritima complexed with 5-formyl-thf
64 c3lx8A_
not modelled
97.7
18
PDB header: metal transportChain: A: PDB Molecule: ferrous iron uptake transporter protein b;PDBTitle: crystal structure of gdp-bound nfeob from s. thermophilus
65 c1s0uA_
not modelled
97.7
13
PDB header: translationChain: A: PDB Molecule: translation initiation factor 2 gamma subunit;PDBTitle: eif2gamma apo
66 d1u94a1
not modelled
97.7
23
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)67 c2gszE_
not modelled
97.7
21
PDB header: protein transportChain: E: PDB Molecule: twitching motility protein pilt;PDBTitle: structure of a. aeolicus pilt with 6 monomers per2 asymmetric unit
68 c1mj1A_
not modelled
97.7
15
PDB header: ribosomeChain: A: PDB Molecule: elongation factor tu;PDBTitle: fitting the ternary complex of ef-tu/trna/gtp and ribosomal proteins2 into a 13 a cryo-em map of the coli 70s ribosome
69 c2qagA_
not modelled
97.7
18
PDB header: cell cycle, structural proteinChain: A: PDB Molecule: septin-2;PDBTitle: crystal structure of human septin trimer 2/6/7
70 d1ubea1
not modelled
97.7
24
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)71 c1udxA_
not modelled
97.7
16
PDB header: protein bindingChain: A: PDB Molecule: the gtp-binding protein obg;PDBTitle: crystal structure of the conserved protein tt1381 from thermus2 thermophilus hb8
72 c1lnzA_
not modelled
97.7
25
PDB header: cell cycleChain: A: PDB Molecule: spo0b-associated gtp-binding protein;PDBTitle: structure of the obg gtp-binding protein
73 c1g7tA_
not modelled
97.7
17
PDB header: translationChain: A: PDB Molecule: translation initiation factor if2/eif5b;PDBTitle: x-ray structure of translation initiation factor if2/eif5b2 complexed with gdpnp
74 d1mo6a1
not modelled
97.7
23
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)75 c3la6P_
not modelled
97.7
19
PDB header: transferaseChain: P: PDB Molecule: tyrosine-protein kinase wzc;PDBTitle: octameric kinase domain of the e. coli tyrosine kinase wzc with bound2 adp
76 c2recB_
not modelled
97.6
23
PDB header: helicasePDB COMPND: 77 c2eyuA_
not modelled
97.6
21
PDB header: protein transportChain: A: PDB Molecule: twitching motility protein pilt;PDBTitle: the crystal structure of the c-terminal domain of aquifex2 aeolicus pilt
78 c2h5eB_
not modelled
97.6
17
PDB header: translationChain: B: PDB Molecule: peptide chain release factor rf-3;PDBTitle: crystal structure of e.coli polypeptide release factor rf3
79 c3ievA_
not modelled
97.6
16
PDB header: nucleotide binding protein/rnaChain: A: PDB Molecule: gtp-binding protein era;PDBTitle: crystal structure of era in complex with mggnp and the 3' end of 16s2 rrna
80 c2ywfA_
not modelled
97.6
19
PDB header: translationChain: A: PDB Molecule: gtp-binding protein lepa;PDBTitle: crystal structure of gmppnp-bound lepa from aquifex aeolicus
81 c2zroA_
not modelled
97.6
24
PDB header: hydrolaseChain: A: PDB Molecule: protein reca;PDBTitle: msreca adp form iv
82 c2elfA_
not modelled
97.6
23
PDB header: translationChain: A: PDB Molecule: protein translation elongation factor 1a;PDBTitle: crystal structure of the selb-like elongation factor ef-pyl2 from methanosarcina mazei
83 c3hr8A_
not modelled
97.6
22
PDB header: recombinationChain: A: PDB Molecule: protein reca;PDBTitle: crystal structure of thermotoga maritima reca
84 c2vyeA_
not modelled
97.6
16
PDB header: hydrolaseChain: A: PDB Molecule: replicative dna helicase;PDBTitle: crystal structure of the dnac-ssdna complex
85 c3cioA_
not modelled
97.6
13
PDB header: signaling protein, transferaseChain: A: PDB Molecule: tyrosine-protein kinase etk;PDBTitle: the kinase domain of escherichia coli tyrosine kinase etk
86 c1zo1I_
not modelled
97.5
12
PDB header: translation/rnaChain: I: PDB Molecule: translation initiation factor 2;PDBTitle: if2, if1, and trna fitted to cryo-em data of e. coli 70s2 initiation complex
87 c3mcaA_
not modelled
97.5
17
PDB header: translation regulation/hydrolaseChain: A: PDB Molecule: elongation factor 1 alpha-like protein;PDBTitle: structure of the dom34-hbs1 complex and implications for its role in2 no-go decay
88 d2fh5b1
not modelled
97.5
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins89 c2hjgA_
not modelled
97.5
12
PDB header: hydrolaseChain: A: PDB Molecule: gtp-binding protein enga;PDBTitle: the crystal structure of the b. subtilis yphc gtpase in2 complex with gdp
90 d1f60a3
not modelled
97.5
16
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins91 d2bv3a2
not modelled
97.5
18
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins92 d1xp8a1
not modelled
97.5
22
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)93 c2vedA_
not modelled
97.5
9
PDB header: transferaseChain: A: PDB Molecule: membrane protein capa1, protein tyrosine kinase;PDBTitle: crystal structure of the chimerical mutant capabk55m2 protein
94 c3p1jC_
not modelled
97.4
10
PDB header: hydrolaseChain: C: PDB Molecule: gtpase imap family member 2;PDBTitle: crystal structure of human gtpase imap family member 2 in the2 nucleotide-free state
95 c1xp8A_
not modelled
97.4
22
PDB header: dna binding proteinChain: A: PDB Molecule: reca protein;PDBTitle: "deinococcus radiodurans reca in complex with atp-gamma-s"
96 c3lxxA_
not modelled
97.4
12
PDB header: immune systemChain: A: PDB Molecule: gtpase imap family member 4;PDBTitle: crystal structure of human gtpase imap family member 4
97 c2q3fB_
not modelled
97.4
16
PDB header: protein bindingChain: B: PDB Molecule: ras-related gtp-binding protein d;PDBTitle: x-ray crystal structure of putative human ras-related gtp2 binding d in complex with gmppnp
98 c3tr5C_
not modelled
97.4
17
PDB header: translationChain: C: PDB Molecule: peptide chain release factor 3;PDBTitle: structure of a peptide chain release factor 3 (prfc) from coxiella2 burnetii
99 d1puia_
not modelled
97.4
18
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins100 c2ja1A_
not modelled
97.4
17
PDB header: transferaseChain: A: PDB Molecule: thymidine kinase;PDBTitle: thymidine kinase from b. cereus with ttp bound as phosphate2 donor.
101 c3lxwA_
not modelled
97.4
12
PDB header: immune systemChain: A: PDB Molecule: gtpase imap family member 1;PDBTitle: crystal structure of human gtpase imap family member 1
102 d1zunb3
not modelled
97.4
16
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins103 c2q6tB_
not modelled
97.3
13
PDB header: hydrolaseChain: B: PDB Molecule: dnab replication fork helicase;PDBTitle: crystal structure of the thermus aquaticus dnab monomer
104 c3r7wC_
not modelled
97.3
15
PDB header: protein transportChain: C: PDB Molecule: gtp-binding protein gtr1;PDBTitle: crystal structure of gtr1p-gtr2p complex
105 c1jalA_
not modelled
97.3
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: ychf protein;PDBTitle: ychf protein (hi0393)
106 c2xexA_
not modelled
97.3
18
PDB header: translationChain: A: PDB Molecule: elongation factor g;PDBTitle: crystal structure of staphylococcus aureus elongation factor2 g
107 c2bm0A_
not modelled
97.3
21
PDB header: elongation factorChain: A: PDB Molecule: elongation factor g;PDBTitle: ribosomal elongation factor g (ef-g) fusidic acid resistant2 mutant t84a
108 d1g3qa_
not modelled
97.3
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like109 c2dy1A_
not modelled
97.3
19
PDB header: signaling protein, translationChain: A: PDB Molecule: elongation factor g;PDBTitle: crystal structure of ef-g-2 from thermus thermophilus
110 c2xtnA_
not modelled
97.3
9
PDB header: immune systemChain: A: PDB Molecule: gtpase imap family member 2;PDBTitle: crystal structure of gtp-bound human gimap2, amino acid2 residues 1-234
111 c1xx6B_
not modelled
97.3
15
PDB header: transferaseChain: B: PDB Molecule: thymidine kinase;PDBTitle: x-ray structure of clostridium acetobutylicum thymidine kinase with2 adp. northeast structural genomics target car26.
112 d1svia_
not modelled
97.3
18
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins113 c3mmpC_
not modelled
97.2
14
PDB header: transferaseChain: C: PDB Molecule: elongation factor tu 2, elongation factor ts;PDBTitle: structure of the qb replicase, an rna-dependent rna polymerase2 consisting of viral and host proteins
114 c1skqB_
not modelled
97.2
17
PDB header: translationChain: B: PDB Molecule: elongation factor 1-alpha;PDBTitle: the crystal structure of sulfolobus solfataricus elongation2 factor 1-alpha in complex with magnesium and gdp
115 d1qhxa_
not modelled
97.2
30
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Chloramphenicol phosphotransferase116 c3degC_
not modelled
97.2
14
PDB header: ribosomeChain: C: PDB Molecule: gtp-binding protein lepa;PDBTitle: complex of elongating escherichia coli 70s ribosome and ef4(lepa)-2 gmppnp
117 c3cmvG_
not modelled
97.2
23
PDB header: recombinationChain: G: PDB Molecule: protein reca;PDBTitle: mechanism of homologous recombination from the reca-2 ssdna/dsdna structures
118 d1tf7a2
not modelled
97.2
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)119 d1ihua2
not modelled
97.2
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like120 c2ztsB_
not modelled
97.2
20
PDB header: atp-binding proteinChain: B: PDB Molecule: putative uncharacterized protein ph0186;PDBTitle: crystal structure of kaic-like protein ph0186 from2 hyperthermophilic archaea pyrococcus horikoshii ot3