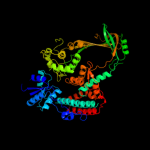
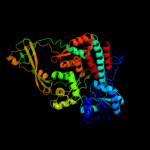
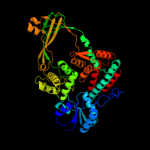
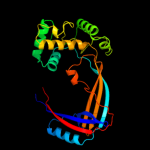
1 d1mw9x_
100.0
98
Fold: Prokaryotic type I DNA topoisomeraseSuperfamily: Prokaryotic type I DNA topoisomeraseFamily: Prokaryotic type I DNA topoisomerase2 c2gajA_
100.0
38
PDB header: isomeraseChain: A: PDB Molecule: dna topoisomerase i;PDBTitle: structure of full length topoisomerase i from thermotoga maritima in2 monoclinic crystal form
3 c2o59B_
100.0
25
PDB header: isomerase/dnaChain: B: PDB Molecule: dna topoisomerase 3;PDBTitle: structure of e. coli topoisomerase iii in complex with an 8-2 base single stranded oligonucleotide. frozen in glycerol3 ph 8.0
4 d1i7da_
100.0
25
Fold: Prokaryotic type I DNA topoisomeraseSuperfamily: Prokaryotic type I DNA topoisomeraseFamily: Prokaryotic type I DNA topoisomerase5 c1gl9B_
100.0
28
PDB header: topoisomeraseChain: B: PDB Molecule: reverse gyrase;PDBTitle: archaeoglobus fulgidus reverse gyrase complexed with adpnp
6 d1gkub3
100.0
28
Fold: Prokaryotic type I DNA topoisomeraseSuperfamily: Prokaryotic type I DNA topoisomeraseFamily: Prokaryotic type I DNA topoisomerase7 d1cy9a_
100.0
98
Fold: Prokaryotic type I DNA topoisomeraseSuperfamily: Prokaryotic type I DNA topoisomeraseFamily: Prokaryotic type I DNA topoisomerase8 c1yuaA_
100.0
99
PDB header: dna binding proteinChain: A: PDB Molecule: topoisomerase i;PDBTitle: c-terminal domain of escherichia coli topoisomerase i
9 d1yuaa2
99.9
100
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: Prokaryotic DNA topoisomerase I, a C-terminal fragment10 d1yuaa1
99.4
98
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: Prokaryotic DNA topoisomerase I, a C-terminal fragment11 d1vdda_
99.2
25
Fold: Recombination protein RecRSuperfamily: Recombination protein RecRFamily: Recombination protein RecR12 c1vddC_
99.0
25
PDB header: recombinationChain: C: PDB Molecule: recombination protein recr;PDBTitle: crystal structure of recombinational repair protein recr
13 d2fcja1
97.0
22
Fold: Toprim domainSuperfamily: Toprim domainFamily: Toprim domain14 c1i3qI_
96.5
13
PDB header: transcriptionChain: I: PDB Molecule: dna-directed rna polymerase ii 14.2kdPDBTitle: rna polymerase ii crystal form i at 3.1 a resolution
15 d1nuia1
95.8
21
Fold: DNA primase coreSuperfamily: DNA primase coreFamily: Primase fragment of primase-helicase protein16 c3h0gI_
95.6
17
PDB header: transcriptionChain: I: PDB Molecule: dna-directed rna polymerase ii subunit rpb9;PDBTitle: rna polymerase ii from schizosaccharomyces pombe
17 c1nuiA_
95.2
21
PDB header: replicationChain: A: PDB Molecule: dna primase/helicase;PDBTitle: crystal structure of the primase fragment of bacteriophage t7 primase-2 helicase protein
18 c2x4hA_
95.0
14
PDB header: transcriptionChain: A: PDB Molecule: hypothetical protein sso2273;PDBTitle: crystal structure of the hypothetical protein sso2273 from2 sulfolobus solfataricus
19 c1q57G_
93.4
24
PDB header: transferaseChain: G: PDB Molecule: dna primase/helicase;PDBTitle: the crystal structure of the bifunctional primase-helicase of2 bacteriophage t7
20 c3g3zA_
92.9
16
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator, marr family;PDBTitle: the structure of nmb1585, a marr family regulator from neisseria2 meningitidis
21 c3hruA_
not modelled
92.0
8
PDB header: transcriptionChain: A: PDB Molecule: metalloregulator scar;PDBTitle: crystal structure of scar with bound zn2+
22 c1g3wA_
not modelled
90.9
9
PDB header: gene regulationChain: A: PDB Molecule: diphtheria toxin repressor;PDBTitle: cd-cys102ser dtxr
23 c1f5tA_
not modelled
90.5
9
PDB header: transcription/dnaChain: A: PDB Molecule: diphtheria toxin repressor;PDBTitle: diphtheria tox repressor (c102d mutant) complexed with2 nickel and dtxr consensus binding sequence
24 d2fxaa1
not modelled
90.5
28
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators25 d2fbia1
not modelled
90.1
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators26 c3nrvC_
not modelled
90.0
12
PDB header: transcription regulatorChain: C: PDB Molecule: putative transcriptional regulator (marr/emrr family);PDBTitle: crystal structure of marr/emrr family transcriptional regulator from2 acinetobacter sp. adp1
27 d2fiya1
not modelled
89.9
18
Fold: FdhE-likeSuperfamily: FdhE-likeFamily: FdhE-like28 d1pvma3
not modelled
89.7
29
Fold: Rubredoxin-likeSuperfamily: Hypothetical protein Ta0289 C-terminal domainFamily: Hypothetical protein Ta0289 C-terminal domain29 c2pexA_
not modelled
89.4
13
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator ohrr;PDBTitle: structure of reduced c22s ohrr from xanthamonas campestris
30 c1fx7C_
not modelled
88.9
9
PDB header: signaling proteinChain: C: PDB Molecule: iron-dependent repressor ider;PDBTitle: crystal structure of the iron-dependent regulator (ider)2 from mycobacterium tuberculosis
31 c3s2wB_
not modelled
88.7
15
PDB header: transcription regulatorChain: B: PDB Molecule: transcriptional regulator, marr family;PDBTitle: the crystal structure of a marr transcriptional regulator from2 methanosarcina mazei go1
32 d1t6t1_
not modelled
88.6
24
Fold: Toprim domainSuperfamily: Toprim domainFamily: Toprim domain33 d2a61a1
not modelled
88.4
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators34 d1lnwa_
not modelled
88.3
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators35 d1dgsa1
not modelled
88.1
36
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: NAD+-dependent DNA ligase, domain 336 c3boqB_
not modelled
88.1
7
PDB header: transcription regulatorChain: B: PDB Molecule: transcriptional regulator, marr family;PDBTitle: crystal structure of marr family transcriptional regulator from2 silicibacter pomeroyi
37 c3bjaA_
not modelled
87.6
17
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator, marr family, putative;PDBTitle: crystal structure of putative marr-like transcription regulator2 (np_978771.1) from bacillus cereus atcc 10987 at 2.38 a resolution
38 d1lj9a_
not modelled
87.3
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators39 c3jw4C_
not modelled
87.2
13
PDB header: transcription regulatorChain: C: PDB Molecule: transcriptional regulator, marr/emrr family;PDBTitle: the structure of a putative marr family transcriptional regulator from2 clostridium acetobutylicum
40 c2hr5B_
not modelled
87.2
25
PDB header: metal binding proteinChain: B: PDB Molecule: rubrerythrin;PDBTitle: pf1283- rubrerythrin from pyrococcus furiosus iron bound form
41 c2gxgA_
not modelled
86.9
11
PDB header: transcriptionChain: A: PDB Molecule: 146aa long hypothetical transcriptional regulator;PDBTitle: crystal structure of emrr homolog from hyperthermophilic archaea2 sulfolobus tokodaii strain7
42 d1s3ja_
not modelled
86.9
10
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators43 c3bj6B_
not modelled
86.8
14
PDB header: transcription regulatorChain: B: PDB Molecule: transcriptional regulator, marr family;PDBTitle: crystal structure of marr family transcription regulator sp03579
44 d3ctaa1
not modelled
86.5
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators45 c2fxaB_
not modelled
86.4
27
PDB header: transcriptionChain: B: PDB Molecule: protease production regulatory protein hpr;PDBTitle: structure of the protease production regulatory protein hpr from2 bacillus subtilis.
46 c2h09A_
not modelled
86.4
19
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator mntr;PDBTitle: crystal structure of diphtheria toxin repressor like protein2 from e. coli
47 c1v9pB_
not modelled
86.4
33
PDB header: ligaseChain: B: PDB Molecule: dna ligase;PDBTitle: crystal structure of nad+-dependent dna ligase
48 c2f5qA_
not modelled
86.3
29
PDB header: hydrolase/dnaChain: A: PDB Molecule: formamidopyrimidine-dna glycosidase;PDBTitle: catalytically inactive (e3q) mutm crosslinked to oxog:c2 containing dna cc2
49 c3kp3B_
not modelled
86.3
5
PDB header: transcription regulator/antibioticChain: B: PDB Molecule: transcriptional regulator tcar;PDBTitle: staphylococcus epidermidis in complex with ampicillin
50 c2qwwB_
not modelled
86.1
17
PDB header: transcriptionChain: B: PDB Molecule: transcriptional regulator, marr family;PDBTitle: crystal structure of multiple antibiotic-resistance repressor (marr)2 (yp_013417.1) from listeria monocytogenes 4b f2365 at 2.07 a3 resolution
51 c3bpxB_
not modelled
86.0
16
PDB header: transcription regulatorChain: B: PDB Molecule: transcriptional regulator;PDBTitle: crystal structure of marr
52 c1dgsB_
not modelled
85.9
33
PDB header: ligaseChain: B: PDB Molecule: dna ligase;PDBTitle: crystal structure of nad+-dependent dna ligase from t.2 filiformis
53 c2opfA_
not modelled
85.4
19
PDB header: hydrolase/dnaChain: A: PDB Molecule: endonuclease viii;PDBTitle: crystal structure of the dna repair enzyme endonuclease-viii (nei)2 from e. coli (r252a) in complex with ap-site containing dna substrate
54 c2fa5B_
not modelled
85.3
16
PDB header: transcriptionChain: B: PDB Molecule: transcriptional regulator marr/emrr family;PDBTitle: the crystal structure of an unliganded multiple antibiotic-2 resistance repressor (marr) from xanthomonas campestris
55 c2it0A_
not modelled
85.3
9
PDB header: transcription/dnaChain: A: PDB Molecule: iron-dependent repressor ider;PDBTitle: crystal structure of a two-domain ider-dna complex crystal2 form ii
56 d1twfi2
not modelled
85.2
14
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: Transcriptional factor domain57 c1k82D_
not modelled
85.1
29
PDB header: hydrolase/dnaChain: D: PDB Molecule: formamidopyrimidine-dna glycosylase;PDBTitle: crystal structure of e.coli formamidopyrimidine-dna2 glycosylase (fpg) covalently trapped with dna
58 c1nnjA_
not modelled
85.0
19
PDB header: hydrolaseChain: A: PDB Molecule: formamidopyrimidine-dna glycosylase;PDBTitle: crystal structure complex between the lactococcus lactis fpg and an2 abasic site containing dna
59 c2rdpA_
not modelled
84.8
21
PDB header: transcriptionChain: A: PDB Molecule: putative transcriptional regulator marr;PDBTitle: the structure of a marr family protein from bacillus2 stearothermophilus
60 d2hr3a1
not modelled
84.6
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators61 d2bv6a1
not modelled
84.5
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators62 d1p4xa1
not modelled
84.4
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators63 c2nyxB_
not modelled
84.3
14
PDB header: transcriptionChain: B: PDB Molecule: probable transcriptional regulatory protein, rv1404;PDBTitle: crystal structure of rv1404 from mycobacterium tuberculosis
64 c3floD_
not modelled
84.3
20
PDB header: transferaseChain: D: PDB Molecule: dna polymerase alpha catalytic subunit a;PDBTitle: crystal structure of the carboxyl-terminal domain of yeast2 dna polymerase alpha in complex with its b subunit
65 d2fbha1
not modelled
84.3
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators66 c1yuzB_
not modelled
84.2
18
PDB header: oxidoreductaseChain: B: PDB Molecule: nigerythrin;PDBTitle: partially reduced state of nigerythrin
67 c1ee8A_
not modelled
84.0
28
PDB header: dna binding proteinChain: A: PDB Molecule: mutm (fpg) protein;PDBTitle: crystal structure of mutm (fpg) protein from thermus thermophilus hb8
68 c3on1A_
not modelled
83.9
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: bh2414 protein;PDBTitle: the structure of a protein with unknown function from bacillus2 halodurans c
69 c3e6mD_
not modelled
83.6
13
PDB header: transcription regulatorChain: D: PDB Molecule: marr family transcriptional regulator;PDBTitle: the crystal structure of a marr family transcriptional2 regulator from silicibacter pomeroyi dss.
70 c3f3xA_
not modelled
83.6
12
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator, marr family, putative;PDBTitle: crystal structure of the transcriptional regulator bldr2 from sulfolobus solfataricus
71 d3broa1
not modelled
83.4
6
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators72 d1r7ja_
not modelled
83.2
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Archaeal DNA-binding protein73 d1jgsa_
not modelled
83.1
7
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators74 c3nqoB_
not modelled
83.1
13
PDB header: transcriptionChain: B: PDB Molecule: marr-family transcriptional regulator;PDBTitle: crystal structure of a marr family transcriptional regulator (cd1569)2 from clostridium difficile 630 at 2.20 a resolution
75 c3cdhB_
not modelled
82.6
24
PDB header: transcription regulatorChain: B: PDB Molecule: transcriptional regulator, marr family;PDBTitle: crystal structure of the marr family transcriptional regulator spo14532 from silicibacter pomeroyi dss-3
76 d2etha1
not modelled
82.2
20
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators77 d1p4xa2
not modelled
81.8
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators78 c2owoA_
not modelled
81.7
32
PDB header: ligase/dnaChain: A: PDB Molecule: dna ligase;PDBTitle: last stop on the road to repair: structure of e.coli dna ligase bound2 to nicked dna-adenylate
79 d1stza1
not modelled
81.0
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Heat-inducible transcription repressor HrcA, N-terminal domain80 c3fm5D_
not modelled
80.5
19
PDB header: transcription regulatorChain: D: PDB Molecule: transcriptional regulator;PDBTitle: x-ray crystal structure of transcriptional regulator (marr family)2 from rhodococcus sp. rha1
81 c3cjnA_
not modelled
79.9
25
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator, marr family;PDBTitle: crystal structure of transcriptional regulator, marr family, from2 silicibacter pomeroyi
82 c2nnnB_
not modelled
79.4
13
PDB header: transcriptionChain: B: PDB Molecule: probable transcriptional regulator;PDBTitle: crystal structure of probable transcriptional regulator from2 pseudomonas aeruginosa
83 c3hrmA_
not modelled
79.4
14
PDB header: transcription regulatorChain: A: PDB Molecule: hth-type transcriptional regulator sarz;PDBTitle: crystal structure of staphylococcus aureus protein sarz in sulfenic2 acid form
84 c3k0lA_
not modelled
79.3
11
PDB header: transcription regulatorChain: A: PDB Molecule: repressor protein;PDBTitle: crystal structure of putative marr family transcriptional2 regulator from acinetobacter sp. adp
85 c1p4xA_
not modelled
78.8
13
PDB header: transcriptionChain: A: PDB Molecule: staphylococcal accessory regulator a homologue;PDBTitle: crystal structure of sars protein from staphylococcus aureus
86 d1ee8a3
not modelled
78.0
28
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: C-terminal, Zn-finger domain of MutM-like DNA repair proteins87 d1ub9a_
not modelled
76.9
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators88 d1tdza3
not modelled
76.6
19
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: C-terminal, Zn-finger domain of MutM-like DNA repair proteins89 c3f2cA_
not modelled
76.5
31
PDB header: transferase/dnaChain: A: PDB Molecule: geobacillus kaustophilus dna polc;PDBTitle: dna polymerase polc from geobacillus kaustophilus complex with dna,2 dgtp and mn
90 d1l1ta3
not modelled
76.0
29
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: C-terminal, Zn-finger domain of MutM-like DNA repair proteins91 d1r2za3
not modelled
75.4
29
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: C-terminal, Zn-finger domain of MutM-like DNA repair proteins92 d1k82a3
not modelled
74.9
29
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: C-terminal, Zn-finger domain of MutM-like DNA repair proteins93 c3ecoB_
not modelled
74.7
10
PDB header: transcriptionChain: B: PDB Molecule: mepr;PDBTitle: crystal structure of mepr, a transcription regulator of the2 staphylococcus aureus multidrug efflux pump mepa
94 c2jrpA_
not modelled
73.9
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative cytoplasmic protein;PDBTitle: solution nmr structure of yfgj from salmonella typhimurium2 modeled with two zn+2 bound, northeast structural genomics3 consortium target str86
95 c2au3A_
not modelled
73.8
22
PDB header: transferaseChain: A: PDB Molecule: dna primase;PDBTitle: crystal structure of the aquifex aeolicus primase (zinc binding and2 rna polymerase domains)
96 c3oopA_
not modelled
73.3
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: lin2960 protein;PDBTitle: the structure of a protein with unknown function from listeria innocua2 clip11262
97 c3deuB_
not modelled
73.0
10
PDB header: transcription regulatorChain: B: PDB Molecule: transcriptional regulator slya;PDBTitle: crystal structure of transcription regulatory protein slya2 from salmonella typhimurium in complex with salicylate3 ligands
98 d1k3xa3
not modelled
73.0
21
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: C-terminal, Zn-finger domain of MutM-like DNA repair proteins99 d2fbka1
not modelled
72.9
10
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators100 d3deua1
not modelled
72.6
8
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators101 c3tgnA_
not modelled
72.4
11
PDB header: transcriptionChain: A: PDB Molecule: adc operon repressor adcr;PDBTitle: crystal structure of the zinc-dependent marr family transcriptional2 regulator adcr in the zn(ii)-bound state
102 c3cngC_
not modelled
71.6
23
PDB header: hydrolaseChain: C: PDB Molecule: nudix hydrolase;PDBTitle: crystal structure of nudix hydrolase from nitrosomonas europaea
103 c1dvbA_
not modelled
71.0
24
PDB header: electron transportChain: A: PDB Molecule: rubrerythrin;PDBTitle: rubrerythrin
104 c2ev5B_
not modelled
70.7
14
PDB header: transcriptionChain: B: PDB Molecule: transcriptional regulator mntr;PDBTitle: bacillus subtilis manganese transport regulator (mntr)2 bound to calcium
105 c3nuhB_
not modelled
70.7
19
PDB header: isomeraseChain: B: PDB Molecule: dna gyrase subunit b;PDBTitle: a domain insertion in e. coli gyrb adopts a novel fold that plays a2 critical role in gyrase function
106 c2wteB_
not modelled
70.5
15
PDB header: antiviral proteinChain: B: PDB Molecule: csa3;PDBTitle: the structure of the crispr-associated protein, csa3, from2 sulfolobus solfataricus at 1.8 angstrom resolution.
107 d1olta_
not modelled
69.7
13
Fold: TIM beta/alpha-barrelSuperfamily: Radical SAM enzymesFamily: Oxygen-independent coproporphyrinogen III oxidase HemN108 c3ri2B_
not modelled
68.7
23
PDB header: transcription regulatorChain: B: PDB Molecule: transcriptional regulator, padr-like family;PDBTitle: crystal structure of padr family transcriptional regulator from2 eggerthella lenta dsm 2243
109 c2hu9B_
not modelled
67.4
25
PDB header: metal transportChain: B: PDB Molecule: mercuric transport protein periplasmic component;PDBTitle: x-ray structure of the archaeoglobus fulgidus copz n-2 terminal domain
110 d1qypa_
not modelled
67.2
33
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: Transcriptional factor domain111 d1z91a1
not modelled
66.4
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators112 d2isya1
not modelled
65.8
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Iron-dependent repressor protein113 c3l09B_
not modelled
65.0
17
PDB header: transcription regulatorChain: B: PDB Molecule: putative transcriptional regulator;PDBTitle: crystal structure of putative transcriptional regulator2 (jann_22dec04_contig27_revised_gene3569) from jannaschia sp. ccs1 at3 2.81 a resolution
114 c3ndjA_
not modelled
64.4
18
PDB header: transferaseChain: A: PDB Molecule: methyltransferase;PDBTitle: x-ray structure of a c-3'-methyltransferase in complex with s-2 adenosyl-l-homocysteine and sugar product
115 c2jneA_
not modelled
63.6
16
PDB header: metal binding proteinChain: A: PDB Molecule: hypothetical protein yfgj;PDBTitle: nmr structure of e.coli yfgj modelled with two zn+2 bound.2 northeast structural genomics consortium target er317.
116 d2jnea1
not modelled
63.6
16
Fold: Rubredoxin-likeSuperfamily: YfgJ-likeFamily: YfgJ-like117 c2k4bA_
not modelled
63.4
7
PDB header: dna binding proteinChain: A: PDB Molecule: transcriptional regulator;PDBTitle: copr repressor structure
118 d1g3wa1
not modelled
61.1
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Iron-dependent repressor protein119 d1jhfa1
not modelled
60.7
9
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: LexA repressor, N-terminal DNA-binding domain120 d2p6ra1
not modelled
59.7
23
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: RecQ helicase DNA-binding domain-like