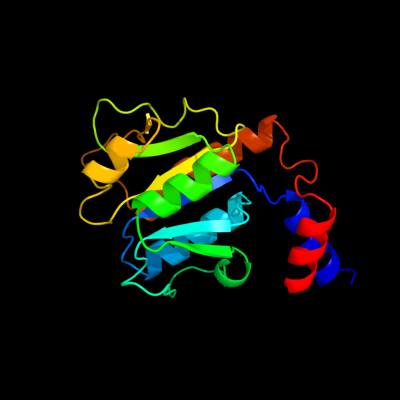
| 1 | c1zjrA_
|
|
 |
100.0 |
42 |
PDB header:transferase
Chain: A: PDB Molecule:trna (guanosine-2'-o-)-methyltransferase;
PDBTitle: crystal structure of a. aeolicus trmh/spou trna modifying enzyme
|
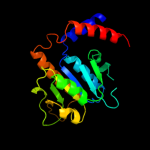
| 2 | d1v2xa_
|
|
 |
100.0 |
53 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:SpoU-like RNA 2'-O ribose methyltransferase |
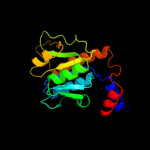
| 3 | d1gz0a1
|
|
 |
100.0 |
28 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:SpoU-like RNA 2'-O ribose methyltransferase |
| 4 | c1gz0G_
|
|
 |
100.0 |
28 |
PDB header:transferase
Chain: G: PDB Molecule:hypothetical trna/rrna methyltransferase yjfh;
PDBTitle: 23s ribosomal rna g2251 2'o-methyltransferase rlmb
|
| 5 | c2ha8A_
|
|
 |
100.0 |
22 |
PDB header:rna binding protein
Chain: A: PDB Molecule:tar (hiv-1) rna loop binding protein;
PDBTitle: methyltransferase domain of human tar (hiv-1) rna binding2 protein 1
|
| 6 | c1x7pB_
|
|
 |
100.0 |
29 |
PDB header:transferase
Chain: B: PDB Molecule:rrna methyltransferase;
PDBTitle: crystal structure of the spou methyltransferase avirb from2 streptomyces viridochromogenes in complex with the cofactor adomet
|
| 7 | c1ipaA_
|
|
 |
100.0 |
28 |
PDB header:transferase
Chain: A: PDB Molecule:rna 2'-o-ribose methyltransferase;
PDBTitle: crystal structure of rna 2'-o ribose methyltransferase
|
| 8 | c2i6dA_
|
|
 |
100.0 |
28 |
PDB header:transferase
Chain: A: PDB Molecule:rna methyltransferase, trmh family;
PDBTitle: the structure of a putative rna methyltransferase of the trmh family2 from porphyromonas gingivalis.
|
| 9 | c3gyqB_
|
|
 |
100.0 |
24 |
PDB header:transferase
Chain: B: PDB Molecule:rrna (adenosine-2'-o-)-methyltransferase;
PDBTitle: structure of the thiostrepton-resistance methyltransferase2 s-adenosyl-l-methionine complex
|
| 10 | d1ipaa1
|
|
 |
100.0 |
27 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:SpoU-like RNA 2'-O ribose methyltransferase |
| 11 | c3ic6A_
|
|
 |
100.0 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative methylase family protein;
PDBTitle: crystal structure of putative methylase family protein from neisseria2 gonorrhoeae
|
| 12 | d1mxia_
|
|
 |
100.0 |
23 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:SpoU-like RNA 2'-O ribose methyltransferase |
| 13 | c3ilkB_
|
|
 |
100.0 |
18 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized trna/rrna methyltransferase hi0380;
PDBTitle: the structure of a probable methylase family protein from haemophilus2 influenzae rd kw20
|
| 14 | c3onpA_
|
|
 |
100.0 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:trna/rrna methyltransferase (spou);
PDBTitle: crystal structure of trna/rrna methyltransferase spou from rhodobacter2 sphaeroides
|
| 15 | c3e5yB_
|
|
 |
100.0 |
24 |
PDB header:transferase
Chain: B: PDB Molecule:trmh family rna methyltransferase;
PDBTitle: crystal structure of trmh family rna methyltransferase from2 burkholderia pseudomallei
|
| 16 | c3l8uA_
|
|
 |
100.0 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:putative rrna methylase;
PDBTitle: crystal structure of smu.1707c, a putative rrna methyltransferase from2 streptococcus mutans ua159
|
| 17 | c3ktyA_
|
|
 |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:probable methyltransferase;
PDBTitle: crystal structure of probable methyltransferase from bordetella2 pertussis tohama i
|
| 18 | c3dcmX_
|
|
 |
99.9 |
18 |
PDB header:transferase
Chain: X: PDB Molecule:uncharacterized protein tm_1570;
PDBTitle: crystal structure of the thermotoga maritima spout family2 rna-methyltransferase protein tm1570 in complex with s-3 adenosyl-l-methionine
|
| 19 | d1vhka2
|
|
 |
97.4 |
15 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:YggJ C-terminal domain-like |
| 20 | c1vhkA_
|
|
 |
97.0 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein yqeu;
PDBTitle: crystal structure of an hypothetical protein
|
| 21 | c2yy8B_ |
|
not modelled |
96.6 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:upf0106 protein ph0461;
PDBTitle: crystal structure of archaeal trna-methylase for position2 56 (atrm56) from pyrococcus horikoshii, complexed with s-3 adenosyl-l-methionine
|
| 22 | c3kw2A_ |
|
not modelled |
96.4 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:probable r-rna methyltransferase;
PDBTitle: crystal structure of probable rrna-methyltransferase from2 porphyromonas gingivalis
|
| 23 | c2egwB_ |
|
not modelled |
96.3 |
10 |
PDB header:rna methyltransferase
Chain: B: PDB Molecule:upf0088 protein aq_165;
PDBTitle: crystal structure of rrna methyltransferase with sah ligand
|
| 24 | c3ai9X_ |
|
not modelled |
96.2 |
16 |
PDB header:transferase
Chain: X: PDB Molecule:upf0217 protein mj1640;
PDBTitle: crystal structure of duf358 protein reveals a putative spout-class2 rrna methyltransferase
|
| 25 | d1nxza2 |
|
not modelled |
96.2 |
10 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:YggJ C-terminal domain-like |
| 26 | d2o3aa1 |
|
not modelled |
95.4 |
18 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:AF0751-like |
| 27 | c1vhyB_ |
|
not modelled |
94.7 |
14 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical protein hi0303;
PDBTitle: crystal structure of haemophilus influenzae protein hi0303, pfam2 duf558
|
| 28 | d2qwva1 |
|
not modelled |
94.3 |
15 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:AF1056-like |
| 29 | d1v6za2 |
|
not modelled |
92.3 |
21 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:YggJ C-terminal domain-like |
| 30 | d2qmma1 |
|
not modelled |
88.1 |
14 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:AF1056-like |
| 31 | c2cx8B_ |
|
not modelled |
86.9 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:methyl transferase;
PDBTitle: crystal structure of methyltransferase with ligand(sah)
|
| 32 | c1z85B_ |
|
not modelled |
60.9 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:hypothetical protein tm1380;
PDBTitle: crystal structure of a predicted rna methyltransferase (tm1380) from2 thermotoga maritima msb8 at 2.12 a resolution
|
| 33 | c1k3rA_ |
|
not modelled |
49.6 |
25 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:conserved protein mt0001;
PDBTitle: crystal structure of the methyltransferase with a knot from2 methanobacterium thermoautotrophicum
|
| 34 | d1ml4a2 |
|
not modelled |
47.2 |
13 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 35 | d1a9xa3 |
|
not modelled |
40.3 |
12 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 36 | c2v3jA_ |
|
not modelled |
39.2 |
16 |
PDB header:ribosomal protein
Chain: A: PDB Molecule:essential for mitotic growth 1;
PDBTitle: the yeast ribosome synthesis factor emg1 alpha beta knot2 fold methyltransferase
|
| 37 | d2b7oa1 |
|
not modelled |
38.7 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class-II DAHP synthetase |
| 38 | d1o6da_ |
|
not modelled |
38.6 |
26 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:YbeA-like |
| 39 | d1j6ua2 |
|
not modelled |
31.8 |
11 |
Fold:MurD-like peptide ligases, peptide-binding domain
Superfamily:MurD-like peptide ligases, peptide-binding domain
Family:MurCDEF C-terminal domain |
| 40 | d1to0a_ |
|
not modelled |
31.0 |
14 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:YbeA-like |
| 41 | d1k3ra2 |
|
not modelled |
30.7 |
26 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:Hypothetical protein MTH1 (MT0001), dimerisation domain |
| 42 | c1j6uA_ |
|
not modelled |
30.0 |
13 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramate-alanine ligase murc;
PDBTitle: crystal structure of udp-n-acetylmuramate-alanine ligase2 murc (tm0231) from thermotoga maritima at 2.3 a resolution
|
| 43 | d1vh0a_ |
|
not modelled |
29.8 |
11 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:YbeA-like |
| 44 | d1ns5a_ |
|
not modelled |
29.6 |
26 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:YbeA-like |
| 45 | d1mx3a1 |
|
not modelled |
29.3 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 46 | d1ekxa2 |
|
not modelled |
28.9 |
15 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 47 | c2omeA_ |
|
not modelled |
27.0 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:c-terminal-binding protein 2;
PDBTitle: crystal structure of human ctbp2 dehydrogenase complexed with nad(h)
|
| 48 | d1qpoa1 |
|
not modelled |
25.7 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:Nicotinate/Quinolinate PRTase C-terminal domain-like
Family:NadC C-terminal domain-like |
| 49 | c2wl8D_ |
|
not modelled |
25.7 |
14 |
PDB header:protein transport
Chain: D: PDB Molecule:peroxisomal biogenesis factor 19;
PDBTitle: x-ray crystal structure of pex19p
|
| 50 | d1o4ua1 |
|
not modelled |
25.1 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Nicotinate/Quinolinate PRTase C-terminal domain-like
Family:NadC C-terminal domain-like |
| 51 | c3gg9C_ |
|
not modelled |
24.8 |
6 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:d-3-phosphoglycerate dehydrogenase oxidoreductase protein;
PDBTitle: crystal structure of putative d-3-phosphoglycerate dehydrogenase2 oxidoreductase from ralstonia solanacearum
|
| 52 | c3o7bA_ |
|
not modelled |
23.2 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:ribosome biogenesis nep1 rna methyltransferase;
PDBTitle: crystal structure of archaeoglobus fulgidus nep1 bound to s-2 adenosylhomocysteine
|
| 53 | c3czpA_ |
|
not modelled |
21.2 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:putative polyphosphate kinase 2;
PDBTitle: crystal structure of putative polyphosphate kinase 2 from pseudomonas2 aeruginosa pa01
|
| 54 | c1qpoA_ |
|
not modelled |
20.4 |
9 |
PDB header:transferase
Chain: A: PDB Molecule:quinolinate acid phosphoribosyl transferase;
PDBTitle: quinolinate phosphoribosyl transferase (qaprtase) apo-enzyme from2 mycobacterium tuberculosis
|
| 55 | c2y0fD_ |
|
not modelled |
19.9 |
29 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase;
PDBTitle: structure of gcpe (ispg) from thermus thermophilus hb27
|
| 56 | c2o8vA_ |
|
not modelled |
19.8 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:phosphoadenosine phosphosulfate reductase;
PDBTitle: paps reductase in a covalent complex with thioredoxin c35a
|
| 57 | c2oq2B_ |
|
not modelled |
19.0 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:phosphoadenosine phosphosulfate reductase;
PDBTitle: crystal structure of yeast paps reductase with pap, a product complex
|
| 58 | d1sura_ |
|
not modelled |
18.1 |
15 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:PAPS reductase-like |
| 59 | c2x3yA_ |
|
not modelled |
18.0 |
19 |
PDB header:isomerase
Chain: A: PDB Molecule:phosphoheptose isomerase;
PDBTitle: crystal structure of gmha from burkholderia pseudomallei
|
| 60 | c2yqeA_ |
|
not modelled |
17.7 |
15 |
PDB header:dna binding protein
Chain: A: PDB Molecule:jumonji/arid domain-containing protein 1d;
PDBTitle: solution structure of the arid domain of jarid1d protein
|
| 61 | d1nkua_ |
|
not modelled |
17.3 |
29 |
Fold:DNA-glycosylase
Superfamily:DNA-glycosylase
Family:3-Methyladenine DNA glycosylase I (Tag) |
| 62 | c2jg6A_ |
|
not modelled |
16.8 |
25 |
PDB header:hydrolase
Chain: A: PDB Molecule:dna-3-methyladenine glycosidase;
PDBTitle: crystal structure of a 3-methyladenine dna glycosylase i2 from staphylococcus aureus
|
| 63 | c3ajfA_ |
|
not modelled |
16.6 |
25 |
PDB header:viral protein
Chain: A: PDB Molecule:non-structural protein 3;
PDBTitle: structural insigths into dsrna binding and rna silencing suppression2 by ns3 protein of rice hoja blanca tenuivirus
|
| 64 | c2zy3A_ |
|
not modelled |
16.4 |
26 |
PDB header:lyase
Chain: A: PDB Molecule:l-aspartate beta-decarboxylase;
PDBTitle: dodecameric l-aspartate beta-decarboxylase
|
| 65 | c2w2kB_ |
|
not modelled |
16.3 |
10 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:d-mandelate dehydrogenase;
PDBTitle: crystal structure of the apo forms of rhodotorula graminis2 d-mandelate dehydrogenase at 1.8a.
|
| 66 | c3q4gA_ |
|
not modelled |
16.2 |
17 |
PDB header:ligase
Chain: A: PDB Molecule:nh(3)-dependent nad(+) synthetase;
PDBTitle: structure of nad synthetase from vibrio cholerae
|
| 67 | c1dvpA_ |
|
not modelled |
15.3 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:hepatocyte growth factor-regulated tyrosine
PDBTitle: crystal structure of the vhs and fyve tandem domains of hrs,2 a protein involved in membrane trafficking and signal3 transduction
|
| 68 | c2fe3B_ |
|
not modelled |
15.1 |
19 |
PDB header:dna binding protein
Chain: B: PDB Molecule:peroxide operon regulator;
PDBTitle: the crystal structure of bacillus subtilis perr-zn reveals a novel2 zn(cys)4 structural redox switch
|
| 69 | c2b7pA_ |
|
not modelled |
14.9 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:probable nicotinate-nucleotide pyrophosphorylase;
PDBTitle: crystal structure of quinolinic acid phosphoribosyltransferase from2 helicobacter pylori
|
| 70 | d2f06a1 |
|
not modelled |
14.8 |
17 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:BT0572-like |
| 71 | d2v3ka1 |
|
not modelled |
14.5 |
17 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:EMG1/NEP1-like |
| 72 | c2jbmA_ |
|
not modelled |
14.5 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:nicotinate-nucleotide pyrophosphorylase;
PDBTitle: qprtase structure from human
|
| 73 | c3mwmA_ |
|
not modelled |
14.4 |
13 |
PDB header:transcription
Chain: A: PDB Molecule:putative metal uptake regulation protein;
PDBTitle: graded expression of zinc-responsive genes through two regulatory2 zinc-binding sites in zur
|
| 74 | d1e8ca2 |
|
not modelled |
14.4 |
8 |
Fold:MurD-like peptide ligases, peptide-binding domain
Superfamily:MurD-like peptide ligases, peptide-binding domain
Family:MurCDEF C-terminal domain |
| 75 | d1x7da_ |
|
not modelled |
13.4 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Ornithine cyclodeaminase-like |
| 76 | c2qipA_ |
|
not modelled |
13.4 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:protein of unknown function vpa0982;
PDBTitle: crystal structure of a protein of unknown function vpa0982 from vibrio2 parahaemolyticus rimd 2210633
|
| 77 | d1o17a2 |
|
not modelled |
12.8 |
45 |
Fold:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Superfamily:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Family:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain |
| 78 | c2o03A_ |
|
not modelled |
12.6 |
15 |
PDB header:gene regulation
Chain: A: PDB Molecule:probable zinc uptake regulation protein furb;
PDBTitle: crystal structure of furb from m. tuberculosis- a zinc uptake2 regulator
|
| 79 | c3fxaA_ |
|
not modelled |
12.5 |
12 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:sis domain protein;
PDBTitle: crystal structure of a putative sugar-phosphate isomerase2 (lmof2365_0531) from listeria monocytogenes str. 4b f2365 at 1.60 a3 resolution
|
| 80 | c2goyC_ |
|
not modelled |
12.4 |
19 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:adenosine phosphosulfate reductase;
PDBTitle: crystal structure of assimilatory adenosine 5'-2 phosphosulfate reductase with bound aps
|
| 81 | d2gy9b1 |
|
not modelled |
12.3 |
23 |
Fold:Flavodoxin-like
Superfamily:Ribosomal protein S2
Family:Ribosomal protein S2 |
| 82 | d1dvpa1 |
|
not modelled |
12.1 |
19 |
Fold:alpha-alpha superhelix
Superfamily:ENTH/VHS domain
Family:VHS domain |
| 83 | c2nacA_ |
|
not modelled |
11.8 |
12 |
PDB header:oxidoreductase(aldehyde(d),nad+(a))
Chain: A: PDB Molecule:nad-dependent formate dehydrogenase;
PDBTitle: high resolution structures of holo and apo formate dehydrogenase
|
| 84 | d1pg5a2 |
|
not modelled |
11.3 |
13 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 85 | c2zkqb_ |
|
not modelled |
11.2 |
18 |
PDB header:ribosomal protein/rna
Chain: B: PDB Molecule:rna expansion segment es3;
PDBTitle: structure of a mammalian ribosomal 40s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
|
| 86 | c3navB_ |
|
not modelled |
10.8 |
7 |
PDB header:lyase
Chain: B: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of an alpha subunit of tryptophan synthase from2 vibrio cholerae o1 biovar el tor str. n16961
|
| 87 | d1q9ua_ |
|
not modelled |
10.4 |
21 |
Fold:TBP-like
Superfamily:TT1751-like
Family:TT1751-like |
| 88 | c1x3lA_ |
|
not modelled |
10.3 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:hypothetical protein ph0495;
PDBTitle: crystal structure of the ph0495 protein from pyrococccus horikoshii2 ot3
|
| 89 | d1oaoc_ |
|
not modelled |
10.0 |
15 |
Fold:Prismane protein-like
Superfamily:Prismane protein-like
Family:Acetyl-CoA synthase |
| 90 | c1o4uA_ |
|
not modelled |
9.2 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:type ii quinolic acid phosphoribosyltransferase;
PDBTitle: crystal structure of a nicotinate nucleotide pyrophosphorylase2 (tm1645) from thermotoga maritima at 2.50 a resolution
|
| 91 | d1kkxa_ |
|
not modelled |
9.2 |
14 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:ARID-like
Family:ARID domain |
| 92 | c3hpbA_ |
|
not modelled |
9.2 |
23 |
PDB header:protein transport
Chain: A: PDB Molecule:snx5 protein;
PDBTitle: crystal structure of snx5-px domain in p212121 space group
|
| 93 | c3bchA_ |
|
not modelled |
9.1 |
21 |
PDB header:cell adhesion, ribosomal protein
Chain: A: PDB Molecule:40s ribosomal protein sa;
PDBTitle: crystal structure of the human laminin receptor precursor
|
| 94 | d1duvg2 |
|
not modelled |
9.1 |
10 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 95 | d1hjra_ |
|
not modelled |
8.9 |
33 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:RuvC resolvase |
| 96 | d1dxya1 |
|
not modelled |
8.9 |
11 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 97 | c2kwuA_ |
|
not modelled |
8.7 |
40 |
PDB header:protein binding/signaling protein
Chain: A: PDB Molecule:dna polymerase iota;
PDBTitle: solution structure of ubm2 of murine polymerase iota in complex with2 ubiquitin
|
| 98 | c3l0gD_ |
|
not modelled |
8.7 |
11 |
PDB header:transferase
Chain: D: PDB Molecule:nicotinate-nucleotide pyrophosphorylase;
PDBTitle: crystal structure of nicotinate-nucleotide pyrophosphorylase from2 ehrlichia chaffeensis at 2.05a resolution
|
| 99 | c3bbnB_ |
|
not modelled |
8.7 |
13 |
PDB header:ribosome
Chain: B: PDB Molecule:ribosomal protein s2;
PDBTitle: homology model for the spinach chloroplast 30s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome.
|