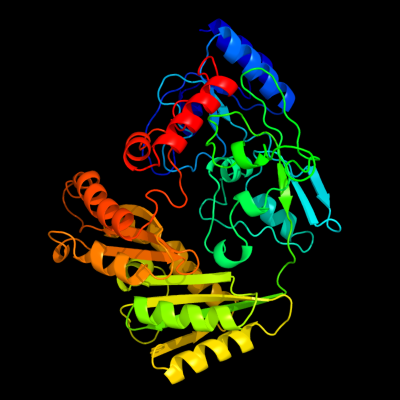
| 1 | c3pm9A_
|
|
 |
100.0 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of a putative dehydrogenase (rpa1076) from2 rhodopseudomonas palustris cga009 at 2.57 a resolution
|
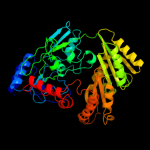
| 2 | c1ahuB_
|
|
 |
100.0 |
18 |
PDB header:flavoenzyme
Chain: B: PDB Molecule:vanillyl-alcohol oxidase;
PDBTitle: structure of the octameric flavoenzyme vanillyl-alcohol2 oxidase in complex with p-cresol
|
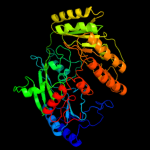
| 3 | c2uuvC_
|
|
 |
100.0 |
21 |
PDB header:transferase
Chain: C: PDB Molecule:alkyldihydroxyacetonephosphate synthase;
PDBTitle: alkyldihydroxyacetonephosphate synthase in p1
|
| 4 | c1f0xA_
|
|
 |
100.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-lactate dehydrogenase;
PDBTitle: crystal structure of d-lactate dehydrogenase, a peripheral2 membrane respiratory enzyme.
|
| 5 | c1wveB_
|
|
 |
100.0 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:4-cresol dehydrogenase [hydroxylating]
PDBTitle: p-cresol methylhydroxylase: alteration of the structure of2 the flavoprotein subunit upon its binding to the3 cytochrome subunit
|
| 6 | c3bw7A_
|
|
 |
100.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytokinin dehydrogenase 1;
PDBTitle: maize cytokinin oxidase/dehydrogenase complexed with the allenic2 cytokinin analog ha-1
|
| 7 | c2exrA_
|
|
 |
100.0 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytokinin dehydrogenase 7;
PDBTitle: x-ray structure of cytokinin oxidase/dehydrogenase (ckx)2 from arabidopsis thaliana at5g21482
|
| 8 | c2bvfA_
|
|
 |
100.0 |
16 |
PDB header:oxidase
Chain: A: PDB Molecule:6-hydroxy-d-nicotine oxidase;
PDBTitle: crystal structure of 6-hydoxy-d-nicotine oxidase from2 arthrobacter nicotinovorans. crystal form 3 (p1)
|
| 9 | c3fwaA_
|
|
 |
100.0 |
13 |
PDB header:flavoprotein
Chain: A: PDB Molecule:reticuline oxidase;
PDBTitle: structure of berberine bridge enzyme, c166a variant in complex with2 (s)-reticuline
|
| 10 | c1zr6A_
|
|
 |
100.0 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glucooligosaccharide oxidase;
PDBTitle: the crystal structure of an acremonium strictum glucooligosaccharide2 oxidase reveals a novel flavinylation
|
| 11 | c3d2hA_
|
|
 |
100.0 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:berberine bridge-forming enzyme;
PDBTitle: structure of berberine bridge enzyme from eschscholzia californica,2 monoclinic crystal form
|
| 12 | c2ipiD_
|
|
 |
100.0 |
13 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:aclacinomycin oxidoreductase (aknox);
PDBTitle: crystal structure of aclacinomycin oxidoreductase
|
| 13 | c3popD_
|
|
 |
100.0 |
17 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:gilr oxidase;
PDBTitle: the crystal structure of gilr, an oxidoreductase that catalyzes the2 terminal step of gilvocarcin biosynthesis
|
| 14 | c2wdwB_
|
|
 |
100.0 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative hexose oxidase;
PDBTitle: the native crystal structure of the primary hexose oxidase (2 dbv29) in antibiotic a40926 biosynthesis
|
| 15 | c2y3rC_
|
|
 |
100.0 |
15 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:taml;
PDBTitle: structure of the tirandamycin-bound fad-dependent2 tirandamycin oxidase taml in p21 space group
|
| 16 | c2vfvA_
|
|
 |
100.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:xylitol oxidase;
PDBTitle: alditol oxidase from streptomyces coelicolor a3(2): complex2 with sulphite
|
| 17 | c1i19B_
|
|
 |
100.0 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:cholesterol oxidase;
PDBTitle: crystal structure of cholesterol oxidase from b.sterolicum
|
| 18 | c3js8A_
|
|
 |
100.0 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cholesterol oxidase;
PDBTitle: solvent-stable cholesterol oxidase
|
| 19 | d1wvfa2
|
|
 |
100.0 |
22 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:FAD-linked oxidases, N-terminal domain |
| 20 | d1f0xa2
|
|
 |
100.0 |
22 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:FAD-linked oxidases, N-terminal domain |
| 21 | d1e8ga2 |
|
not modelled |
100.0 |
21 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:FAD-linked oxidases, N-terminal domain |
| 22 | d1w1oa2 |
|
not modelled |
100.0 |
20 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:FAD-linked oxidases, N-terminal domain |
| 23 | d2i0ka2 |
|
not modelled |
100.0 |
22 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:FAD-linked oxidases, N-terminal domain |
| 24 | d1hska1 |
|
not modelled |
100.0 |
19 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:Uridine diphospho-N-Acetylenolpyruvylglucosamine reductase (MurB), N-terminal domain |
| 25 | d1e8ga1 |
|
not modelled |
100.0 |
16 |
Fold:Ferredoxin-like
Superfamily:FAD-linked oxidases, C-terminal domain
Family:Vanillyl-alcohol oxidase-like |
| 26 | d1uxya1 |
|
not modelled |
100.0 |
12 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:Uridine diphospho-N-Acetylenolpyruvylglucosamine reductase (MurB), N-terminal domain |
| 27 | c1hskA_ |
|
not modelled |
100.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:udp-n-acetylenolpyruvoylglucosamine reductase;
PDBTitle: crystal structure of s. aureus murb
|
| 28 | d1wvfa1 |
|
not modelled |
100.0 |
20 |
Fold:Ferredoxin-like
Superfamily:FAD-linked oxidases, C-terminal domain
Family:Vanillyl-alcohol oxidase-like |
| 29 | d1f0xa1 |
|
not modelled |
100.0 |
20 |
Fold:Ferredoxin-like
Superfamily:FAD-linked oxidases, C-terminal domain
Family:D-lactate dehydrogenase |
| 30 | c2yvsA_ |
|
not modelled |
100.0 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glycolate oxidase subunit glce;
PDBTitle: crystal structure of glycolate oxidase subunit glce from thermus2 thermophilus hb8
|
| 31 | c1mbbA_ |
|
not modelled |
100.0 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:uridine diphospho-n-acetylenolpyruvylglucosamine
PDBTitle: oxidoreductase
|
| 32 | c3i99A_ |
|
not modelled |
99.9 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:udp-n-acetylenolpyruvoylglucosamine reductase;
PDBTitle: the crystal structure of the udp-n-acetylenolpyruvoylglucosamine2 reductase from the vibrio cholerae o1 biovar tor
|
| 33 | c2gquA_ |
|
not modelled |
99.9 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:udp-n-acetylenolpyruvylglucosamine reductase;
PDBTitle: crystal structure of udp-n-acetylenolpyruvylglucosamine2 reductase (murb) from thermus caldophilus
|
| 34 | d1w1oa1 |
|
not modelled |
99.3 |
16 |
Fold:Ferredoxin-like
Superfamily:FAD-linked oxidases, C-terminal domain
Family:Cytokinin dehydrogenase 1 |
| 35 | d1ffvc2 |
|
not modelled |
97.8 |
19 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CO dehydrogenase flavoprotein N-terminal domain-like |
| 36 | c1n62C_ |
|
not modelled |
97.8 |
17 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:carbon monoxide dehydrogenase medium chain;
PDBTitle: crystal structure of the mo,cu-co dehydrogenase (codh), n-2 butylisocyanide-bound state
|
| 37 | c1ffuF_ |
|
not modelled |
97.7 |
20 |
PDB header:hydrolase
Chain: F: PDB Molecule:cutm, flavoprotein of carbon monoxide
PDBTitle: carbon monoxide dehydrogenase from hydrogenophaga2 pseudoflava which lacks the mo-pyranopterin moiety of the3 molybdenum cofactor
|
| 38 | c1t3qF_ |
|
not modelled |
97.5 |
14 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:quinoline 2-oxidoreductase medium subunit;
PDBTitle: crystal structure of quinoline 2-oxidoreductase from pseudomonas2 putida 86
|
| 39 | d3b9jb2 |
|
not modelled |
97.5 |
10 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CO dehydrogenase flavoprotein N-terminal domain-like |
| 40 | c3etrM_ |
|
not modelled |
97.5 |
8 |
PDB header:oxidoreductase
Chain: M: PDB Molecule:xanthine dehydrogenase/oxidase;
PDBTitle: crystal structure of xanthine oxidase in complex with2 lumazine
|
| 41 | c3b9jJ_ |
|
not modelled |
97.5 |
8 |
PDB header:oxidoreductase
Chain: J: PDB Molecule:xanthine oxidase;
PDBTitle: structure of xanthine oxidase with 2-hydroxy-6-methylpurine
|
| 42 | c3hrdC_ |
|
not modelled |
97.4 |
17 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:nicotinate dehydrogenase fad-subunit;
PDBTitle: crystal structure of nicotinate dehydrogenase
|
| 43 | d1v97a6 |
|
not modelled |
97.4 |
10 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CO dehydrogenase flavoprotein N-terminal domain-like |
| 44 | d1t3qc2 |
|
not modelled |
97.3 |
15 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CO dehydrogenase flavoprotein N-terminal domain-like |
| 45 | d1n62c2 |
|
not modelled |
97.3 |
18 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CO dehydrogenase flavoprotein N-terminal domain-like |
| 46 | d1jroa4 |
|
not modelled |
97.2 |
13 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CO dehydrogenase flavoprotein N-terminal domain-like |
| 47 | c1rm6E_ |
|
not modelled |
97.0 |
20 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:4-hydroxybenzoyl-coa reductase beta subunit;
PDBTitle: structure of 4-hydroxybenzoyl-coa reductase from thauera2 aromatica
|
| 48 | c2w3rG_ |
|
not modelled |
96.9 |
16 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:xanthine dehydrogenase;
PDBTitle: crystal structure of xanthine dehydrogenase (desulfo form)2 from rhodobacter capsulatus in complex with hypoxanthine
|
| 49 | d1rm6b2 |
|
not modelled |
96.3 |
21 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CO dehydrogenase flavoprotein N-terminal domain-like |
| 50 | c1wygA_ |
|
not modelled |
95.9 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:xanthine dehydrogenase/oxidase;
PDBTitle: crystal structure of a rat xanthine dehydrogenase triple mutant2 (c535a, c992r and c1324s)
|
| 51 | d2i0ka1 |
|
not modelled |
86.0 |
13 |
Fold:Ferredoxin-like
Superfamily:FAD-linked oxidases, C-terminal domain
Family:Cholesterol oxidase |
| 52 | d1jaka1 |
|
not modelled |
67.9 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-N-acetylhexosaminidase catalytic domain |
| 53 | d1yhta1 |
|
not modelled |
64.9 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-N-acetylhexosaminidase catalytic domain |
| 54 | d1qbaa3 |
|
not modelled |
64.2 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-N-acetylhexosaminidase catalytic domain |
| 55 | d1nowa1 |
|
not modelled |
63.7 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-N-acetylhexosaminidase catalytic domain |
| 56 | c3nsnA_ |
|
not modelled |
63.2 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:n-acetylglucosaminidase;
PDBTitle: crystal structure of insect beta-n-acetyl-d-hexosaminidase ofhex12 complexed with tmg-chitotriomycin
|
| 57 | c1nouA_ |
|
not modelled |
63.1 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-hexosaminidase beta chain;
PDBTitle: native human lysosomal beta-hexosaminidase isoform b
|
| 58 | d2gjxa1 |
|
not modelled |
63.0 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-N-acetylhexosaminidase catalytic domain |
| 59 | c3gh7A_ |
|
not modelled |
62.8 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-hexosaminidase;
PDBTitle: crystal structure of beta-hexosaminidase from paenibacillus2 sp. ts12 in complex with galnac
|
| 60 | c1m04A_ |
|
not modelled |
61.9 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-n-acetylhexosaminidase;
PDBTitle: mutant streptomyces plicatus beta-hexosaminidase (d313n) in complex2 with product (glcnac)
|
| 61 | c3rcnA_ |
|
not modelled |
61.5 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-n-acetylhexosaminidase;
PDBTitle: crystal structure of beta-n-acetylhexosaminidase from arthrobacter2 aurescens
|
| 62 | c2yl8A_ |
|
not modelled |
61.4 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-n-acetylhexosaminidase;
PDBTitle: inhibition of the pneumococcal virulence factor strh and2 molecular insights into n-glycan recognition and3 hydrolysis
|
| 63 | c2uvaI_ |
|
not modelled |
61.3 |
17 |
PDB header:transferase
Chain: I: PDB Molecule:fatty acid synthase beta subunits;
PDBTitle: crystal structure of fatty acid synthase from thermomyces2 lanuginosus at 3.1 angstrom resolution. this file contains3 the beta subunits of the fatty acid synthase. the entire4 crystal structure consists of one heterododecameric fatty5 acid synthase and is described in remark 400
|
| 64 | c2ylaA_ |
|
not modelled |
61.1 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-n-acetylhexosaminidase;
PDBTitle: inhibition of the pneumococcal virulence factor strh and2 molecular insights into n-glycan recognition and3 hydrolysis
|
| 65 | c2dgxA_ |
|
not modelled |
59.9 |
16 |
PDB header:rna binding protein
Chain: A: PDB Molecule:kiaa0430 protein;
PDBTitle: solution structure of the rna recognition motif in kiaa04302 protein
|
| 66 | c2gjxE_ |
|
not modelled |
59.7 |
10 |
PDB header:hydrolase
Chain: E: PDB Molecule:beta-hexosaminidase alpha chain;
PDBTitle: crystallographic structure of human beta-hexosaminidase a
|
| 67 | c3lmyA_ |
|
not modelled |
59.6 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-hexosaminidase subunit beta;
PDBTitle: the crystal structure of beta-hexosaminidase b in complex with2 pyrimethamine
|
| 68 | c1qupA_ |
|
not modelled |
59.1 |
11 |
PDB header:chaperone
Chain: A: PDB Molecule:superoxide dismutase 1 copper chaperone;
PDBTitle: crystal structure of the copper chaperone for superoxide2 dismutase
|
| 69 | c2vkzH_ |
|
not modelled |
58.9 |
19 |
PDB header:transferase
Chain: H: PDB Molecule:fatty acid synthase subunit beta;
PDBTitle: structure of the cerulenin-inhibited fungal fatty acid2 synthase type i multienzyme complex
|
| 70 | c3rpmA_ |
|
not modelled |
56.5 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-n-acetyl-hexosaminidase;
PDBTitle: crystal structure of the first gh20 domain of a novel beta-n-acetyl-2 hexosaminidase strh from streptococcus pneumoniae r6
|
| 71 | c1qbaA_ |
|
not modelled |
52.4 |
10 |
PDB header:glycosyl hydrolase
Chain: A: PDB Molecule:chitobiase;
PDBTitle: bacterial chitobiase, glycosyl hydrolase family 20
|
| 72 | c1jk9D_ |
|
not modelled |
51.1 |
11 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:copper chaperone for superoxide dismutase;
PDBTitle: heterodimer between h48f-ysod1 and yccs
|
| 73 | c2epoB_ |
|
not modelled |
50.1 |
10 |
PDB header:hydrolase
Chain: B: PDB Molecule:n-acetyl-beta-d-glucosaminidase;
PDBTitle: n-acetyl-b-d-glucosaminidase (gcna) from streptococcus gordonii
|
| 74 | c2bp7F_ |
|
not modelled |
46.6 |
10 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:2-oxoisovalerate dehydrogenase beta subunit;
PDBTitle: new crystal form of the pseudomonas putida branched-chain2 dehydrogenase (e1)
|
| 75 | d2p0la1 |
|
not modelled |
45.9 |
16 |
Fold:Class II aaRS and biotin synthetases
Superfamily:Class II aaRS and biotin synthetases
Family:LplA-like |
| 76 | d2p5ia1 |
|
not modelled |
44.1 |
33 |
Fold:Class II aaRS and biotin synthetases
Superfamily:Class II aaRS and biotin synthetases
Family:LplA-like |
| 77 | c2yswB_ |
|
not modelled |
39.2 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: crystal structure of the 3-dehydroquinate dehydratase from aquifex2 aeolicus vf5
|
| 78 | d2bi7a1 |
|
not modelled |
39.1 |
19 |
Fold:Nucleotide-binding domain
Superfamily:Nucleotide-binding domain
Family:UDP-galactopyranose mutase, N-terminal domain |
| 79 | d2cqpa1 |
|
not modelled |
38.5 |
19 |
Fold:Ferredoxin-like
Superfamily:RNA-binding domain, RBD
Family:Canonical RBD |
| 80 | c3s01A_ |
|
not modelled |
38.0 |
12 |
PDB header:rna binding protein
Chain: A: PDB Molecule:heterogeneous nuclear ribonucleoprotein l;
PDBTitle: crystal structure of a heterogeneous nuclear ribonucleoprotein l2 (hnrpl) from mus musculus at 2.15 a resolution
|
| 81 | c2do0A_ |
|
not modelled |
37.4 |
16 |
PDB header:rna binding protein
Chain: A: PDB Molecule:heterogeneous nuclear ribonucleoprotein m;
PDBTitle: solution structure of the rna binding domain of2 heterogeneous nuclear ribonucleoprotein m
|
| 82 | d1a9xb2 |
|
not modelled |
36.8 |
17 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 83 | c2x24B_ |
|
not modelled |
36.6 |
22 |
PDB header:ligase
Chain: B: PDB Molecule:acetyl-coa carboxylase;
PDBTitle: bovine acc2 ct domain in complex with inhibitor
|
| 84 | d1itza2 |
|
not modelled |
35.5 |
11 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:TK-like Pyr module |
| 85 | c3ff6D_ |
|
not modelled |
34.3 |
22 |
PDB header:ligase
Chain: D: PDB Molecule:acetyl-coa carboxylase 2;
PDBTitle: human acc2 ct domain with cp-640186
|
| 86 | d1wexa_ |
|
not modelled |
33.5 |
17 |
Fold:Ferredoxin-like
Superfamily:RNA-binding domain, RBD
Family:Canonical RBD |
| 87 | d1umdb1 |
|
not modelled |
33.3 |
26 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Branched-chain alpha-keto acid dehydrogenase Pyr module |
| 88 | d2r8oa1 |
|
not modelled |
32.8 |
11 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:TK-like Pyr module |
| 89 | c1od4C_ |
|
not modelled |
32.7 |
19 |
PDB header:ligase
Chain: C: PDB Molecule:acetyl-coenzyme a carboxylase;
PDBTitle: acetyl-coa carboxylase carboxyltransferase domain
|
| 90 | d2plsa1 |
|
not modelled |
32.4 |
7 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CorC/HlyC domain-like |
| 91 | c2f3jA_ |
|
not modelled |
31.2 |
16 |
PDB header:transport protein
Chain: A: PDB Molecule:rna and export factor binding protein 2;
PDBTitle: the solution structure of the ref2-i mrna export factor2 (residues 1-155).
|
| 92 | d1no8a_ |
|
not modelled |
29.8 |
16 |
Fold:Ferredoxin-like
Superfamily:RNA-binding domain, RBD
Family:Canonical RBD |
| 93 | c1no8A_ |
|
not modelled |
29.8 |
16 |
PDB header:rna binding protein
Chain: A: PDB Molecule:aly;
PDBTitle: solution structure of the nuclear factor aly rbd domain
|
| 94 | d1qupa2 |
|
not modelled |
29.6 |
14 |
Fold:Ferredoxin-like
Superfamily:HMA, heavy metal-associated domain
Family:HMA, heavy metal-associated domain |
| 95 | c2dnoA_ |
|
not modelled |
29.3 |
21 |
PDB header:rna binding protein
Chain: A: PDB Molecule:trinucleotide repeat containing 4 variant;
PDBTitle: solution structure of rna binding domain in trinucleotide2 repeat containing 4 variant
|
| 96 | c2e44A_ |
|
not modelled |
28.8 |
15 |
PDB header:rna binding protein
Chain: A: PDB Molecule:insulin-like growth factor 2 mrna binding
PDBTitle: solution structure of rna binding domain in insulin-like2 growth factor 2 mrna binding protein 3
|
| 97 | d1r9ja1 |
|
not modelled |
28.3 |
22 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:TK-like Pyr module |
| 98 | c2fy1A_ |
|
not modelled |
28.2 |
21 |
PDB header:structural protein/rna
Chain: A: PDB Molecule:rna-binding motif protein, y chromosome, family
PDBTitle: a dual mode of rna recognition by the rbmy protein
|
| 99 | d2ozlb1 |
|
not modelled |
27.8 |
30 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Branched-chain alpha-keto acid dehydrogenase Pyr module |
| 100 | d1ik6a1 |
|
not modelled |
27.3 |
30 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Branched-chain alpha-keto acid dehydrogenase Pyr module |
| 101 | d1w85b1 |
|
not modelled |
27.1 |
26 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Branched-chain alpha-keto acid dehydrogenase Pyr module |
| 102 | c2gvsA_ |
|
not modelled |
26.8 |
38 |
PDB header:lipid binding protein
Chain: A: PDB Molecule:chemosensory protein csp-sg4;
PDBTitle: nmr solution structure of cspsg4
|
| 103 | c2rneA_ |
|
not modelled |
26.2 |
17 |
PDB header:rna binding protein
Chain: A: PDB Molecule:tia1 protein;
PDBTitle: solution structure of the second rna recognition motif2 (rrm) of tia-1
|
| 104 | d1fhta_ |
|
not modelled |
26.1 |
9 |
Fold:Ferredoxin-like
Superfamily:RNA-binding domain, RBD
Family:Canonical RBD |
| 105 | c3h0jA_ |
|
not modelled |
25.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:acetyl-coa carboxylase;
PDBTitle: crystal structure of the carboxyltransferase domain of2 acetyl-coenzyme a carboxylase in complex with compound 2
|
| 106 | d1kx9b_ |
|
not modelled |
24.6 |
29 |
Fold:alpha-alpha superhelix
Superfamily:Chemosensory protein Csp2
Family:Chemosensory protein Csp2 |
| 107 | d1n8va_ |
|
not modelled |
22.1 |
25 |
Fold:alpha-alpha superhelix
Superfamily:Chemosensory protein Csp2
Family:Chemosensory protein Csp2 |
| 108 | d1qs0b1 |
|
not modelled |
22.0 |
19 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Branched-chain alpha-keto acid dehydrogenase Pyr module |
| 109 | c2crlA_ |
|
not modelled |
22.0 |
3 |
PDB header:chaperone
Chain: A: PDB Molecule:copper chaperone for superoxide dismutase;
PDBTitle: the apo form of hma domain of copper chaperone for2 superoxide dismutase
|
| 110 | d1x5oa1 |
|
not modelled |
22.0 |
17 |
Fold:Ferredoxin-like
Superfamily:RNA-binding domain, RBD
Family:Canonical RBD |
| 111 | c1uytC_ |
|
not modelled |
21.9 |
19 |
PDB header:transferase
Chain: C: PDB Molecule:acetyl-coa carboxylase;
PDBTitle: acetyl-coa carboxylase carboxyltransferase domain
|
| 112 | d1pixa3 |
|
not modelled |
21.7 |
22 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Biotin dependent carboxylase carboxyltransferase domain |
| 113 | c2i38A_ |
|
not modelled |
21.6 |
22 |
PDB header:rna binding protein/chimera
Chain: A: PDB Molecule:fusion protein consists of immunoglobin g-
PDBTitle: solution structure of the rrm of srp20
|
| 114 | c2rd5D_ |
|
not modelled |
21.5 |
31 |
PDB header:protein binding
Chain: D: PDB Molecule:pii protein;
PDBTitle: structural basis for the regulation of n-acetylglutamate kinase by pii2 in arabidopsis thaliana
|
| 115 | c2do4A_ |
|
not modelled |
21.2 |
11 |
PDB header:immune system
Chain: A: PDB Molecule:squamous cell carcinoma antigen recognized by t-
PDBTitle: solution structure of the rna binding domain of squamous2 cell carcinoma antigen recognized by t cells 3
|
| 116 | d2bfdb1 |
|
not modelled |
20.9 |
26 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Branched-chain alpha-keto acid dehydrogenase Pyr module |
| 117 | c3qxbB_ |
|
not modelled |
20.8 |
7 |
PDB header:isomerase
Chain: B: PDB Molecule:putative xylose isomerase;
PDBTitle: crystal structure of a putative xylose isomerase (yp_426450.1) from2 rhodospirillum rubrum atcc 11170 at 1.90 a resolution
|