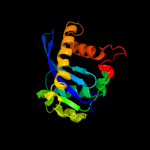
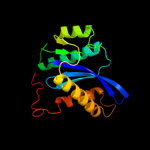
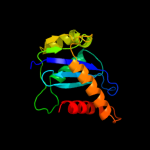
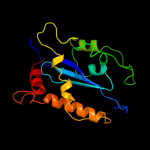
1 c3f9kV_
99.9
18
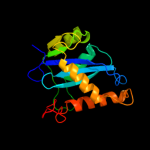
PDB header: viral protein, recombinationChain: V: PDB Molecule: integrase;PDBTitle: two domain fragment of hiv-2 integrase in complex with ledgf ibd
2 d1asua_
99.8
16
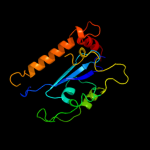
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain3 c1c0mA_
99.8
18
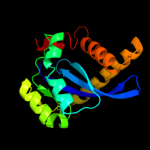
PDB header: transferaseChain: A: PDB Molecule: protein (integrase);PDBTitle: crystal structure of rsv two-domain integrase
4 d1c0ma2
99.8
17
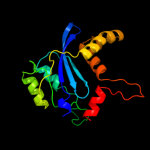
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain5 d1bcoa2
99.8
14
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: mu transposase, core domain6 c3nf9A_
99.8
17
PDB header: hydrolase/hydrolase inhibitorChain: A: PDB Molecule: integrase;PDBTitle: structural basis for a new mechanism of inhibition of hiv integrase2 identified by fragment screening and structure based design
7 d1exqa_
99.8
14
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain8 c1ex4A_
99.8
18
PDB header: viral proteinChain: A: PDB Molecule: integrase;PDBTitle: hiv-1 integrase catalytic core and c-terminal domain
9 c3kksB_
99.7
17
PDB header: dna binding proteinChain: B: PDB Molecule: integrase;PDBTitle: crystal structure of catalytic core domain of biv integrase in crystal2 form ii
10 d1cxqa_
99.7
19
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain11 c1bcoA_
99.7
13
PDB header: transposaseChain: A: PDB Molecule: bacteriophage mu transposase;PDBTitle: bacteriophage mu transposase core domain
12 d1hyva_
99.7
17
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain13 c1k6yB_
99.6
18
PDB header: transferaseChain: B: PDB Molecule: integrase;PDBTitle: crystal structure of a two-domain fragment of hiv-1 integrase
14 c3hpgC_
99.6
21
PDB header: transferaseChain: C: PDB Molecule: integrase;PDBTitle: visna virus integrase (residues 1-219) in complex with ledgf2 ibd: examples of open integrase dimer-dimer interfaces
15 d1c6va_
99.6
19
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain16 c3l2tB_
99.6
15
PDB header: recombination/dnaChain: B: PDB Molecule: integrase;PDBTitle: crystal structure of the prototype foamy virus (pfv) intasome in2 complex with magnesium and mk0518 (raltegravir)
17 c3hosA_
99.5
14
PDB header: transferase, dna binding protein/dnaChain: A: PDB Molecule: transposable element mariner, complete cds;PDBTitle: crystal structure of the mariner mos1 paired end complex with mg
18 c3dlrA_
99.3
14
PDB header: transferaseChain: A: PDB Molecule: integrase;PDBTitle: crystal structure of the catalytic core domain from pfv2 integrase
19 c6paxA_
98.8
17
PDB header: gene regulation/dnaChain: A: PDB Molecule: homeobox protein pax-6;PDBTitle: crystal structure of the human pax-6 paired domain-dna2 complex reveals a general model for pax protein-dna3 interactions
20 d1pdnc_
98.8
20
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Paired domain21 c1u78A_
not modelled
98.6
10
PDB header: dna binding protein/dnaChain: A: PDB Molecule: transposable element tc3 transposase;PDBTitle: structure of the bipartite dna-binding domain of tc32 transposase bound to transposon dna
22 c2k27A_
not modelled
98.4
18
PDB header: transcription regulatorChain: A: PDB Molecule: paired box protein pax-8;PDBTitle: solution structure of human pax8 paired box domain
23 c2f7tA_
not modelled
98.0
14
PDB header: dna binding proteinChain: A: PDB Molecule: mos1 transposase;PDBTitle: crystal structure of the catalytic domain of mos1 mariner2 transposase
24 d2jn6a1
not modelled
97.8
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Cgl2762-like25 c3f2kB_
not modelled
97.2
20
PDB header: transferaseChain: B: PDB Molecule: histone-lysine n-methyltransferase setmar;PDBTitle: structure of the transposase domain of human histone-lysine2 n-methyltransferase setmar
26 c3hefB_
not modelled
97.0
14
PDB header: viral proteinChain: B: PDB Molecule: gene 1 protein;PDBTitle: crystal structure of the bacteriophage sf6 terminase small2 subunit
27 d2coba1
not modelled
96.9
10
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Psq domain28 d1k78a1
not modelled
96.8
26
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Paired domain29 c1hlvA_
not modelled
96.7
11
PDB header: dna binding protein/dnaChain: A: PDB Molecule: major centromere autoantigen b;PDBTitle: crystal structure of cenp-b(1-129) complexed with the cenp-2 b box dna
30 d6paxa1
not modelled
96.7
25
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Paired domain31 c2rn7A_
not modelled
96.6
12
PDB header: unknown functionChain: A: PDB Molecule: is629 orfa;PDBTitle: nmr solution structure of tnpe protein from shigella2 flexneri. northeast structural genomics target sfr125
32 c1umqA_
not modelled
96.4
24
PDB header: dna-binding proteinChain: A: PDB Molecule: photosynthetic apparatus regulatory protein;PDBTitle: solution structure and dna binding of the effector domain2 from the global regulator prra(rega) from r. sphaeroides:3 insights into dna binding specificity
33 d1umqa_
not modelled
96.4
24
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like34 d1ntca_
not modelled
96.3
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like35 c2elhA_
96.1
21
PDB header: dna binding proteinChain: A: PDB Molecule: cg11849-pa;PDBTitle: solution structure of the cenp-b n-terminal dna-binding2 domain of fruit fly distal antenna cg11849-pa
36 d1fipa_
not modelled
96.0
8
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like37 d1etxa_
not modelled
95.9
8
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like38 c3e7lD_
not modelled
95.8
8
PDB header: transcription regulatorChain: D: PDB Molecule: transcriptional regulator (ntrc family);PDBTitle: crystal structure of sigma54 activator ntrc4's dna binding2 domain
39 d1ijwc_
not modelled
95.6
8
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Recombinase DNA-binding domain40 d1g2ha_
not modelled
95.5
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like41 d1hcra_
not modelled
95.5
8
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Recombinase DNA-binding domain42 d1etob_
not modelled
95.3
8
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like43 d1a04a1
not modelled
94.7
24
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: GerE-like (LuxR/UhpA family of transcriptional regulators)44 d2ao9a1
not modelled
94.6
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Nanomeric phage protein-like45 c2w48D_
not modelled
94.5
14
PDB header: transcriptionChain: D: PDB Molecule: sorbitol operon regulator;PDBTitle: crystal structure of the full-length sorbitol operon2 regulator sorc from klebsiella pneumoniae
46 c3fmyA_
not modelled
94.5
6
PDB header: dna binding proteinChain: A: PDB Molecule: hth-type transcriptional regulator mqsaPDBTitle: structure of the c-terminal domain of the e. coli protein2 mqsa (ygit/b3021)
47 c2cg4B_
not modelled
94.2
15
PDB header: transcriptionChain: B: PDB Molecule: regulatory protein asnc;PDBTitle: structure of e.coli asnc
48 c2jvlA_
not modelled
94.0
12
PDB header: transcriptionChain: A: PDB Molecule: trmbf1;PDBTitle: nmr structure of the c-terminal domain of mbf1 of trichoderma reesei
49 c1zljE_
not modelled
93.9
11
PDB header: transcriptionChain: E: PDB Molecule: dormancy survival regulator;PDBTitle: crystal structure of the mycobacterium tuberculosis hypoxic2 response regulator dosr c-terminal domain
50 d1fsea_
not modelled
93.9
10
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: GerE-like (LuxR/UhpA family of transcriptional regulators)51 d1xsva_
not modelled
93.9
20
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: YlxM/p13-like52 d1hlva1
not modelled
93.7
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Centromere-binding53 c2q0oA_
not modelled
93.6
8
PDB header: transcriptionChain: A: PDB Molecule: probable transcriptional activator protein trar;PDBTitle: crystal structure of an anti-activation complex in bacterial quorum2 sensing
54 c3sztB_
not modelled
93.4
8
PDB header: transcriptionChain: B: PDB Molecule: quorum-sensing control repressor;PDBTitle: quorum sensing control repressor, qscr, bound to n-3-oxo-dodecanoyl-l-2 homoserine lactone
55 d1s7oa_
not modelled
93.4
20
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: YlxM/p13-like56 c1r71B_
not modelled
93.4
13
PDB header: transcription/dnaChain: B: PDB Molecule: transcriptional repressor protein korb;PDBTitle: crystal structure of the dna binding domain of korb in2 complex with the operator dna
57 d1yioa1
not modelled
93.3
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: GerE-like (LuxR/UhpA family of transcriptional regulators)58 d2cg4a1
not modelled
93.2
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Lrp/AsnC-like transcriptional regulator N-terminal domain59 d1sq8a_
not modelled
93.1
16
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: Phage repressors60 c3t76A_
not modelled
93.1
16
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator vanug;PDBTitle: crystal structure of transcriptional regulator vanug, form ii
61 d1l3la1
not modelled
92.8
24
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: GerE-like (LuxR/UhpA family of transcriptional regulators)62 d1bw6a_
not modelled
92.7
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Centromere-binding63 c2krfB_
not modelled
92.7
11
PDB header: transcriptionChain: B: PDB Molecule: transcriptional regulatory protein coma;PDBTitle: nmr solution structure of the dna binding domain of competence protein2 a
64 c3hugA_
not modelled
92.6
13
PDB header: transcription/membrane proteinChain: A: PDB Molecule: rna polymerase sigma factor;PDBTitle: crystal structure of mycobacterium tuberculosis anti-sigma factor rsla2 in complex with -35 promoter binding domain of sigl
65 c2l0kA_
not modelled
92.6
10
PDB header: transcriptionChain: A: PDB Molecule: stage iii sporulation protein d;PDBTitle: nmr solution structure of a transcription factor spoiiid in complex2 with dna
66 d1r71a_
not modelled
92.5
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: KorB DNA-binding domain-likeFamily: KorB DNA-binding domain-like67 d1r69a_
not modelled
92.4
15
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: Phage repressors68 d1biaa1
not modelled
92.3
23
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Biotin repressor-like69 c1h0mD_
not modelled
92.3
22
PDB header: transcription/dnaChain: D: PDB Molecule: transcriptional activator protein trar;PDBTitle: three-dimensional structure of the quorum sensing protein2 trar bound to its autoinducer and to its target dna
70 d1vz0a1
not modelled
92.1
23
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: KorB DNA-binding domain-likeFamily: KorB DNA-binding domain-like71 c1x3uA_
not modelled
92.1
11
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulatory protein fixj;PDBTitle: solution structure of the c-terminal transcriptional2 activator domain of fixj from sinorhizobium melilot
72 c2x48B_
not modelled
92.0
28
PDB header: viral proteinChain: B: PDB Molecule: cag38821;PDBTitle: orf 55 from sulfolobus islandicus rudivirus 1
73 d1p4wa_
not modelled
91.8
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: GerE-like (LuxR/UhpA family of transcriptional regulators)74 c3b7hA_
not modelled
91.8
3
PDB header: structural proteinChain: A: PDB Molecule: prophage lp1 protein 11;PDBTitle: crystal structure of the prophage lp1 protein 11
75 c2xcjB_
not modelled
91.6
12
PDB header: viral proteinChain: B: PDB Molecule: c protein;PDBTitle: crystal structure of p2 c, the immunity repressor of2 temperate e. coli phage p2
76 d1y9qa1
not modelled
91.6
26
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: Probable transcriptional regulator VC1968, N-terminal domain77 d1adra_
not modelled
91.6
15
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: Phage repressors78 c3mzyA_
not modelled
91.6
13
PDB header: rna binding proteinChain: A: PDB Molecule: rna polymerase sigma-h factor;PDBTitle: the crystal structure of the rna polymerase sigma-h factor from2 fusobacterium nucleatum to 2.5a
79 c2ia0A_
not modelled
91.6
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative hth-type transcriptional regulator pf0864;PDBTitle: transcriptional regulatory protein pf0864 from pyrococcus furiosus a2 member of the asnc family (pf0864)
80 c2iu5A_
not modelled
91.5
16
PDB header: transcriptionChain: A: PDB Molecule: hth-type dhaklm operon transcriptional activator dhas;PDBTitle: dihydroxyacetone kinase operon activator dhas
81 c3cecA_
not modelled
91.5
18
PDB header: transcriptionChain: A: PDB Molecule: putative antidote protein of plasmid maintenance system;PDBTitle: crystal structure of a putative antidote protein of plasmid2 maintenance system (npun_f2943) from nostoc punctiforme pcc 73102 at3 1.60 a resolution
82 c3qp5C_
not modelled
91.4
11
PDB header: transcriptionChain: C: PDB Molecule: cvir transcriptional regulator;PDBTitle: crystal structure of cvir bound to antagonist chlorolactone (cl)
83 c3o60A_
not modelled
91.4
26
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: lin0861 protein;PDBTitle: the crystal structure of lin0861 from listeria innocua to 2.8a
84 d1j5ya1
not modelled
91.4
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Biotin repressor-like85 c2vbzA_
not modelled
91.4
24
PDB header: dna-binding proteinChain: A: PDB Molecule: transcriptional regulatory protein;PDBTitle: feast or famine regulatory protein (rv3291c)from m.2 tuberculosis complexed with l-tryptophan
86 d2ezla_
not modelled
91.4
21
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Recombinase DNA-binding domain87 c1z4hA_
not modelled
91.3
4
PDB header: protein binding, dna binding proteinChain: A: PDB Molecule: tor inhibition protein;PDBTitle: the response regulator tori belongs to a new family of2 atypical excisionase
88 c2rnjA_
not modelled
91.3
13
PDB header: transcriptionChain: A: PDB Molecule: response regulator protein vrar;PDBTitle: nmr structure of the s. aureus vrar dna binding domain
89 c2w7nA_
not modelled
91.3
10
PDB header: transcription/dnaChain: A: PDB Molecule: trfb transcriptional repressor protein;PDBTitle: crystal structure of kora bound to operator dna: insight2 into repressor cooperation in rp4 gene regulation
90 c2ao9H_
not modelled
91.2
19
PDB header: structural genomics, unknown functionChain: H: PDB Molecule: phage protein;PDBTitle: structural genomics, the crystal structure of a phage protein2 (phbc6a51) from bacillus cereus atcc 14579
91 d1rp3a2
not modelled
91.2
8
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain92 c3op9A_
not modelled
91.2
9
PDB header: transcription regulatorChain: A: PDB Molecule: pli0006 protein;PDBTitle: crystal structure of transcriptional regulator from listeria innocua
93 c2jpcA_
not modelled
91.0
17
PDB header: dna binding proteinChain: A: PDB Molecule: ssrb;PDBTitle: ssrb dna binding protein
94 c3gn5B_
not modelled
90.8
6
PDB header: dna binding proteinChain: B: PDB Molecule: hth-type transcriptional regulator mqsa (ygit/b3021);PDBTitle: structure of the e. coli protein mqsa (ygit/b3021)
95 d1nera_
not modelled
90.8
25
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: Phage repressors96 c2p6tH_
not modelled
90.7
15
PDB header: transcriptionChain: H: PDB Molecule: transcriptional regulator, lrp/asnc family;PDBTitle: crystal structure of transcriptional regulator nmb0573 and l-leucine2 complex from neisseria meningitidis
97 c3c3wB_
not modelled
90.6
8
PDB header: transcriptionChain: B: PDB Molecule: two component transcriptional regulatory protein devr;PDBTitle: crystal structure of the mycobacterium tuberculosis hypoxic response2 regulator dosr
98 c3ivpD_
not modelled
90.5
22
PDB header: dna binding proteinChain: D: PDB Molecule: putative transposon-related dna-binding protein;PDBTitle: the structure of a possible transposon-related dna-binding protein2 from clostridium difficile 630.
99 d1or7a1
not modelled
90.3
5
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain100 d2icta1
not modelled
90.3
12
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: SinR domain-like101 d1gdta1
not modelled
90.2
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Recombinase DNA-binding domain102 c3onqB_
not modelled
90.2
10
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: regulator of polyketide synthase expression;PDBTitle: crystal structure of regulator of polyketide synthase expression2 bad_0249 from bifidobacterium adolescentis
103 c2e7xA_
not modelled
90.2
12
PDB header: transcription regulatorChain: A: PDB Molecule: 150aa long hypothetical transcriptional regulator;PDBTitle: structure of the lrp/asnc like transcriptional regulator from2 sulfolobus tokodaii 7 complexed with its cognate ligand
104 c2np5A_
not modelled
90.2
17
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator;PDBTitle: crystal structure of a transcriptional regulator (rha1_ro04179) from2 rhodococcus sp. rha1.
105 c3f1bA_
not modelled
90.2
15
PDB header: transcription regulatorChain: A: PDB Molecule: tetr-like transcriptional regulator;PDBTitle: the crystal structure of a tetr-like transcriptional regulator from2 rhodococcus sp. rha1.
106 c2fjrB_
not modelled
90.1
11
PDB header: transcription regulatorChain: B: PDB Molecule: repressor protein ci;PDBTitle: crystal structure of bacteriophage 186
107 d1utxa_
not modelled
90.0
20
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: SinR domain-like108 d1uxca_
not modelled
90.0
15
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator109 c1rnlA_
not modelled
90.0
19
PDB header: signal transduction proteinChain: A: PDB Molecule: nitrate/nitrite response regulator protein narl;PDBTitle: the nitrate/nitrite response regulator protein narl from narl
110 c1b0nA_
not modelled
90.0
20
PDB header: transcription regulatorChain: A: PDB Molecule: protein (sinr protein);PDBTitle: sinr protein/sini protein complex
111 c2d6yA_
not modelled
89.9
19
PDB header: gene regulationChain: A: PDB Molecule: putative tetr family regulatory protein;PDBTitle: crystal structure of transcriptional factor sco4008 from streptomyces2 coelicolor a3(2)
112 c3gziA_
not modelled
89.9
13
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator, tetr family;PDBTitle: crystal structure of a transcriptional regulator of the tetr family2 (shew_3567) from shewanella loihica pv-4 at 2.05 a resolution
113 d2fq4a1
not modelled
89.9
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain114 c2hxiA_
not modelled
89.8
25
PDB header: transcriptionChain: A: PDB Molecule: putative transcriptional regulator;PDBTitle: structural genomics, the crystal structure of a putative2 transcriptional regulator from streptomyces coelicolor a3(2)
115 c3eusB_
not modelled
89.7
14
PDB header: dna binding proteinChain: B: PDB Molecule: dna-binding protein;PDBTitle: the crystal structure of the dna binding protein from silicibacter2 pomeroyi
116 d2np5a1
not modelled
89.7
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain117 c1bdhA_
not modelled
89.7
16
PDB header: transcription/dnaChain: A: PDB Molecule: protein (purine repressor);PDBTitle: purine repressor mutant-hypoxanthine-palindromic operator2 complex
118 d1lmb3_
not modelled
89.6
18
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: Phage repressors119 c2e1cA_
not modelled
89.6
15
PDB header: transcription/dnaChain: A: PDB Molecule: putative hth-type transcriptional regulator ph1519;PDBTitle: structure of putative hth-type transcriptional regulator ph1519/dna2 complex
120 c2oqgA_
not modelled
89.6
21
PDB header: transcriptionChain: A: PDB Molecule: possible transcriptional regulator, arsr family protein;PDBTitle: arsr-like transcriptional regulator from rhodococcus sp. rha1