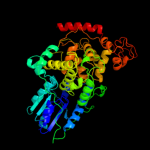
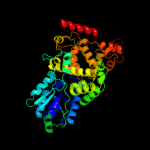
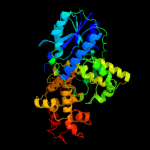
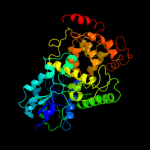
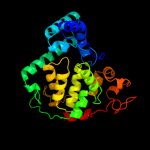
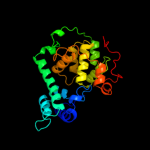
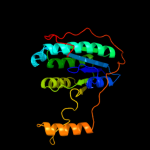
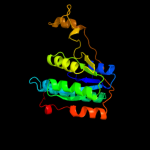
1 c1dnpA_
100.0
100
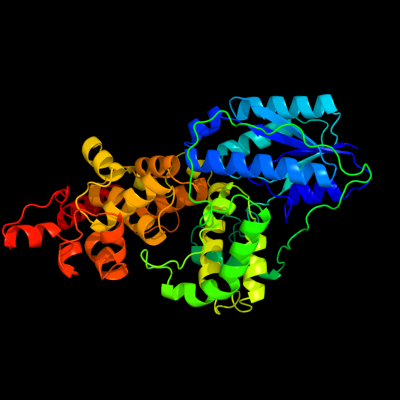
PDB header: lyase (carbon-carbon)Chain: A: PDB Molecule: dna photolyase;PDBTitle: structure of deoxyribodipyrimidine photolyase
2 c3fy4C_
100.0
27
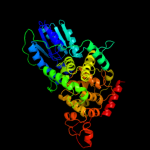
PDB header: lyaseChain: C: PDB Molecule: 6-4 photolyase;PDBTitle: (6-4) photolyase crystal structure
3 c1u3cA_
100.0
29
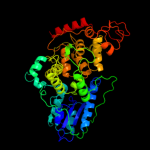
PDB header: signaling proteinChain: A: PDB Molecule: cryptochrome 1 apoprotein;PDBTitle: crystal structure of the phr domain of cryptochrome 1 from2 arabidopsis thaliana
4 c3cvyA_
100.0
28
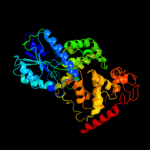
PDB header: lyase/dnaChain: A: PDB Molecule: re11660p;PDBTitle: drosophila melanogaster (6-4) photolyase bound to repaired2 ds dna
5 c1tezB_
100.0
37
PDB header: lyase/dnaChain: B: PDB Molecule: deoxyribodipyrimidine photolyase;PDBTitle: complex between dna and the dna photolyase from anacystis nidulans
6 c1np7A_
100.0
30
PDB header: lyaseChain: A: PDB Molecule: dna photolyase;PDBTitle: crystal structure analysis of synechocystis sp. pcc6803 cryptochrome
7 c3tvsA_
100.0
24
PDB header: signaling proteinChain: A: PDB Molecule: cryptochrome-1;PDBTitle: structure of full-length drosophila cryptochrome
8 c2j4dA_
100.0
26
PDB header: dna-binding proteinChain: A: PDB Molecule: cryptochrome dash;PDBTitle: cryptochrome 3 from arabidopsis thaliana
9 c2e0iD_
100.0
34
PDB header: lyaseChain: D: PDB Molecule: 432aa long hypothetical deoxyribodipyrimidine photolyase;PDBTitle: crystal structure of archaeal photolyase from sulfolobus tokodaii with2 two fad molecules: implication of a novel light-harvesting cofactor
10 c1iqrA_
100.0
36
PDB header: lyaseChain: A: PDB Molecule: photolyase;PDBTitle: crystal structure of dna photolyase from thermus2 thermophilus
11 c2xrzA_
100.0
22
PDB header: lyase/dnaChain: A: PDB Molecule: deoxyribodipyrimidine photolyase;PDBTitle: x-ray structure of archaeal class ii cpd photolyase from2 methanosarcina mazei in complex with intact cpd-lesion
12 c3umvB_
100.0
21
PDB header: lyaseChain: B: PDB Molecule: deoxyribodipyrimidine photo-lyase;PDBTitle: eukaryotic class ii cpd photolyase structure reveals a basis for2 improved uv-tolerance in plants
13 d1dnpa1
100.0
100
Fold: Cryptochrome/photolyase FAD-binding domainSuperfamily: Cryptochrome/photolyase FAD-binding domainFamily: Cryptochrome/photolyase FAD-binding domain14 d1u3da1
100.0
33
Fold: Cryptochrome/photolyase FAD-binding domainSuperfamily: Cryptochrome/photolyase FAD-binding domainFamily: Cryptochrome/photolyase FAD-binding domain15 d1owla1
100.0
48
Fold: Cryptochrome/photolyase FAD-binding domainSuperfamily: Cryptochrome/photolyase FAD-binding domainFamily: Cryptochrome/photolyase FAD-binding domain16 d1np7a1
100.0
34
Fold: Cryptochrome/photolyase FAD-binding domainSuperfamily: Cryptochrome/photolyase FAD-binding domainFamily: Cryptochrome/photolyase FAD-binding domain17 d2j07a1
100.0
45
Fold: Cryptochrome/photolyase FAD-binding domainSuperfamily: Cryptochrome/photolyase FAD-binding domainFamily: Cryptochrome/photolyase FAD-binding domain18 d1dnpa2
100.0
100
Fold: Cryptochrome/photolyase, N-terminal domainSuperfamily: Cryptochrome/photolyase, N-terminal domainFamily: Cryptochrome/photolyase, N-terminal domain19 d1owla2
100.0
23
Fold: Cryptochrome/photolyase, N-terminal domainSuperfamily: Cryptochrome/photolyase, N-terminal domainFamily: Cryptochrome/photolyase, N-terminal domain20 d1u3da2
100.0
24
Fold: Cryptochrome/photolyase, N-terminal domainSuperfamily: Cryptochrome/photolyase, N-terminal domainFamily: Cryptochrome/photolyase, N-terminal domain21 d1np7a2
not modelled
100.0
23
Fold: Cryptochrome/photolyase, N-terminal domainSuperfamily: Cryptochrome/photolyase, N-terminal domainFamily: Cryptochrome/photolyase, N-terminal domain22 d2j07a2
not modelled
100.0
26
Fold: Cryptochrome/photolyase, N-terminal domainSuperfamily: Cryptochrome/photolyase, N-terminal domainFamily: Cryptochrome/photolyase, N-terminal domain23 c2gvsA_
not modelled
75.2
15
PDB header: lipid binding proteinChain: A: PDB Molecule: chemosensory protein csp-sg4;PDBTitle: nmr solution structure of cspsg4
24 d1n8va_
not modelled
72.0
17
Fold: alpha-alpha superhelixSuperfamily: Chemosensory protein Csp2Family: Chemosensory protein Csp225 d1kx9b_
not modelled
68.0
17
Fold: alpha-alpha superhelixSuperfamily: Chemosensory protein Csp2Family: Chemosensory protein Csp226 c1ul1Y_
not modelled
67.8
10
PDB header: hydrolase/dna binding proteinChain: Y: PDB Molecule: flap endonuclease-1;PDBTitle: crystal structure of the human fen1-pcna complex
27 c3obkH_
not modelled
49.4
18
PDB header: lyaseChain: H: PDB Molecule: delta-aminolevulinic acid dehydratase;PDBTitle: crystal structure of delta-aminolevulinic acid dehydratase2 (porphobilinogen synthase) from toxoplasma gondii me49 in complex3 with the reaction product porphobilinogen
28 d1ul1x2
not modelled
32.6
14
Fold: PIN domain-likeSuperfamily: PIN domain-likeFamily: 5' to 3' exonuclease catalytic domain29 c3lgbB_
not modelled
30.6
12
PDB header: transferaseChain: B: PDB Molecule: dna primase large subunit;PDBTitle: crystal structure of the fe-s domain of the yeast dna primase
30 c2l06A_
not modelled
29.3
19
PDB header: protein bindingChain: A: PDB Molecule: phycobilisome lcm core-membrane linker polypeptide;PDBTitle: solution nmr structure of the pbs linker polypeptide domain (fragment2 254-400) of phycobilisome linker protein apce from synechocystis sp.3 pcc 6803. northeast structural genomics consortium target sgr209c
31 c3oryA_
not modelled
27.2
11
PDB header: hydrolaseChain: A: PDB Molecule: flap endonuclease 1;PDBTitle: crystal structure of flap endonuclease 1 from hyperthermophilic2 archaeon desulfurococcus amylolyticus
32 c2pbyB_
not modelled
25.9
12
PDB header: hydrolaseChain: B: PDB Molecule: glutaminase;PDBTitle: probable glutaminase from geobacillus kaustophilus hta426
33 d1l6sa_
not modelled
24.2
17
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: 5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase)34 c2pfsA_
not modelled
23.1
9
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: universal stress protein;PDBTitle: crystal structure of universal stress protein from nitrosomonas2 europaea
35 c2l3wA_
not modelled
22.8
17
PDB header: photosynthesisChain: A: PDB Molecule: phycobilisome rod linker polypeptide;PDBTitle: solution nmr structure of the pbs linker domain of phycobilisome rod2 linker polypeptide from synechococcus elongatus, northeast structural3 genomics consortium target snr168a
36 d1u7ia_
not modelled
22.5
13
Fold: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseSuperfamily: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseFamily: 3-demethylubiquinone-9 3-methyltransferase37 c1a77A_
not modelled
21.3
13
PDB header: 5'-3' exo/endo nucleaseChain: A: PDB Molecule: flap endonuclease-1 protein;PDBTitle: flap endonuclease-1 from methanococcus jannaschii
38 d2d13a1
not modelled
20.0
20
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: N-type ATP pyrophosphatases39 c3mwdA_
not modelled
19.9
13
PDB header: transferaseChain: A: PDB Molecule: atp-citrate synthase;PDBTitle: truncated human atp-citrate lyase with citrate bound
40 c2ky4A_
not modelled
19.9
13
PDB header: photosynthesisChain: A: PDB Molecule: phycobilisome linker polypeptide;PDBTitle: solution nmr structure of the pbs linker domain of phycobilisome2 linker polypeptide from anabaena sp. northeast structural genomics3 consortium target nsr123e
41 d1gzga_
not modelled
19.3
17
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: 5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase)42 c3odhB_
not modelled
17.9
47
PDB header: hydrolase/dnaChain: B: PDB Molecule: okrai endonuclease;PDBTitle: structure of okrai/dna complex
43 d1k8kd2
not modelled
17.8
40
Fold: Secretion chaperone-likeSuperfamily: Arp2/3 complex subunitsFamily: Arp2/3 complex subunits44 c2z86D_
not modelled
17.4
12
PDB header: transferaseChain: D: PDB Molecule: chondroitin synthase;PDBTitle: crystal structure of chondroitin polymerase from2 escherichia coli strain k4 (k4cp) complexed with udp-glcua3 and udp
45 c2izoA_
not modelled
16.7
13
PDB header: hydrolaseChain: A: PDB Molecule: flap structure-specific endonuclease;PDBTitle: structure of an archaeal pcna1-pcna2-fen1 complex
46 c2p9lD_
not modelled
16.4
40
PDB header: structural proteinChain: D: PDB Molecule: actin-related protein 2/3 complex subunit 2;PDBTitle: crystal structure of bovine arp2/3 complex
47 d1b43a2
not modelled
15.1
16
Fold: PIN domain-likeSuperfamily: PIN domain-likeFamily: 5' to 3' exonuclease catalytic domain48 d1zela1
not modelled
14.3
46
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Rv2827c N-terminal domain-like49 c3dwlI_
not modelled
14.2
40
PDB header: structural proteinChain: I: PDB Molecule: actin-related protein 2/3 complex subunit 2;PDBTitle: crystal structure of fission yeast arp2/3 complex lacking the arp22 subunit
50 d2bdua1
not modelled
14.1
16
Fold: HAD-likeSuperfamily: HAD-likeFamily: Pyrimidine 5'-nucleotidase (UMPH-1)51 d2fsqa1
not modelled
13.2
32
Fold: LigT-likeSuperfamily: LigT-likeFamily: Atu0111-like52 d2b8ea1
not modelled
13.1
14
Fold: HAD-likeSuperfamily: HAD-likeFamily: Meta-cation ATPase, catalytic domain P53 d2vkqa1
not modelled
11.4
13
Fold: HAD-likeSuperfamily: HAD-likeFamily: Pyrimidine 5'-nucleotidase (UMPH-1)54 c2qqcC_
not modelled
11.3
13
PDB header: lyaseChain: C: PDB Molecule: pyruvoyl-dependent arginine decarboxylase (ecPDBTitle: e109q mutant of pyruvoyl-dependent arginine decarboxylase2 from methanococcus jannashii
55 c1b43A_
not modelled
10.0
13
PDB header: transferaseChain: A: PDB Molecule: protein (fen-1);PDBTitle: fen-1 from p. furiosus
56 d1nyna_
not modelled
9.2
0
Fold: FYSH domainSuperfamily: FYSH domainFamily: Hypothetical protein Yhr087W57 c3ew8A_
not modelled
9.1
16
PDB header: hydrolaseChain: A: PDB Molecule: histone deacetylase 8;PDBTitle: crystal structure analysis of human hdac8 d101l variant
58 d1t64a_
not modelled
9.0
16
Fold: Arginase/deacetylaseSuperfamily: Arginase/deacetylaseFamily: Histone deacetylase, HDAC59 d1sgma1
not modelled
9.0
23
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain60 d1pv8a_
not modelled
8.4
13
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: 5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase)61 d2ipqx1
not modelled
8.2
6
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: STY4665 C-terminal domain-like62 d2vkva1
not modelled
8.1
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain63 d1evsa_
not modelled
8.0
17
Fold: 4-helical cytokinesSuperfamily: 4-helical cytokinesFamily: Long-chain cytokines64 d1ru8a_
not modelled
7.7
22
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: N-type ATP pyrophosphatases65 d1b74a2
not modelled
7.5
25
Fold: ATC-likeSuperfamily: Aspartate/glutamate racemaseFamily: Aspartate/glutamate racemase66 d2dy1a3
not modelled
7.2
27
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Translational machinery components67 d2bv3a3
not modelled
7.1
50
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Translational machinery components68 d2o7ta1
not modelled
6.8
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain69 c3l9qB_
not modelled
6.7
21
PDB header: transferaseChain: B: PDB Molecule: dna primase large subunit;PDBTitle: crystal structure of human polymerase alpha-primase p58 iron-sulfur2 cluster domain
70 c1p4eB_
not modelled
6.5
30
PDB header: dna binding protein/recombination/dnaChain: B: PDB Molecule: recombinase flp protein;PDBTitle: flpe w330f mutant-dna holliday junction complex
71 c3q8lA_
not modelled
6.2
20
PDB header: hydrolase/dnaChain: A: PDB Molecule: flap endonuclease 1;PDBTitle: crystal structure of human flap endonuclease fen1 (wt) in complex with2 substrate 5'-flap dna, sm3+, and k+
72 c2lmdA_
not modelled
6.2
22
PDB header: transcriptionChain: A: PDB Molecule: prospero homeobox protein 1;PDBTitle: minimal constraints solution nmr structure of prospero homeobox2 protein 1 from homo sapiens, northeast structural genomics consortium3 target hr4660b
73 c2ld7A_
not modelled
6.1
28
PDB header: transcriptionChain: A: PDB Molecule: histone deacetylase complex subunit sap30;PDBTitle: solution structure of the msin3a pah3-sap30 sid complex
74 d1g8fa3
not modelled
6.1
10
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ATP sulfurylase C-terminal domain75 c1rxvA_
not modelled
5.8
11
PDB header: hydrolase/dnaChain: A: PDB Molecule: flap structure-specific endonuclease;PDBTitle: crystal structure of a. fulgidus fen-1 bound to dna
76 c3lklB_
not modelled
5.5
15
PDB header: transport proteinChain: B: PDB Molecule: antisigma-factor antagonist stas;PDBTitle: crystal structure of the c-terminal domain of anti-sigma factor2 antagonist stas from rhodobacter sphaeroides
77 d2i10a1
not modelled
5.5
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain78 c3dloC_
not modelled
5.2
8
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: universal stress protein;PDBTitle: structure of universal stress protein from archaeoglobus fulgidus
79 c3j09A_
not modelled
5.1
13
PDB header: hydrolase, metal transportChain: A: PDB Molecule: copper-exporting p-type atpase a;PDBTitle: high resolution helical reconstruction of the bacterial p-type atpase2 copper transporter copa