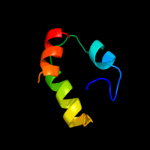
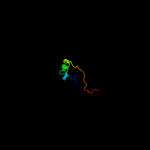
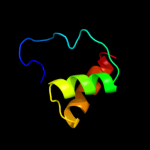
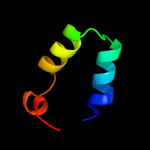
1 c2hl7A_
100.0
44
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome c-type biogenesis protein ccmh;PDBTitle: crystal structure of the periplasmic domain of ccmh from pseudomonas2 aeruginosa
2 c2kw0A_
100.0
43
PDB header: oxidoreductaseChain: A: PDB Molecule: ccmh protein;PDBTitle: solution structure of n-terminal domain of ccmh from escherichia.coli
3 d1pbya1
73.5
15
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Quinohemoprotein amine dehydrogenase A chain, domains 1 and 24 c2qsrA_
66.4
17
PDB header: transcriptionChain: A: PDB Molecule: transcription-repair coupling factor;PDBTitle: crystal structure of c-terminal domain of transcription-repair2 coupling factor
5 d2eyqa6
62.8
37
Fold: TRCF domain-likeSuperfamily: TRCF domain-likeFamily: TRCF domain6 c3omdB_
62.2
9
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of unknown function protein from leptospirillum2 rubarum
7 c2c4rL_
61.0
18
PDB header: hydrolaseChain: L: PDB Molecule: ribonuclease e;PDBTitle: catalytic domain of e. coli rnase e
8 d1r5ya_
59.8
16
Fold: TIM beta/alpha-barrelSuperfamily: tRNA-guanine transglycosylaseFamily: tRNA-guanine transglycosylase9 c2x48B_
54.4
9
PDB header: viral proteinChain: B: PDB Molecule: cag38821;PDBTitle: orf 55 from sulfolobus islandicus rudivirus 1
10 c2bpbB_
54.1
17
PDB header: oxidoreductaseChain: B: PDB Molecule: sulfite\:cytochrome c oxidoreductase subunit b;PDBTitle: sulfite dehydrogenase from starkeya novella
11 c3lm3A_
52.1
27
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a putative glycoside hydrolase/deacetylase2 (bdi_3119) from parabacteroides distasonis at 1.44 a resolution
12 d1jmxa1
46.9
13
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Quinohemoprotein amine dehydrogenase A chain, domains 1 and 213 c3d3mB_
46.4
6
PDB header: translationChain: B: PDB Molecule: eukaryotic translation initiation factor 4 gammaPDBTitle: the crystal structure of the c-terminal region of death2 associated protein 5(dap5)
14 d1fs1b1
42.6
32
Fold: Skp1 dimerisation domain-likeSuperfamily: Skp1 dimerisation domain-likeFamily: Skp1 dimerisation domain-like15 d1fs2b1
40.6
32
Fold: Skp1 dimerisation domain-likeSuperfamily: Skp1 dimerisation domain-likeFamily: Skp1 dimerisation domain-like16 d1ug3a2
39.1
11
Fold: alpha-alpha superhelixSuperfamily: ARM repeatFamily: MIF4G domain-like17 c1iq8B_
38.6
38
PDB header: transferaseChain: B: PDB Molecule: archaeosine trna-guanine transglycosylase;PDBTitle: crystal structure of archaeosine trna-guanine2 transglycosylase from pyrococcus horikoshii
18 d1iq8a1
38.2
20
Fold: TIM beta/alpha-barrelSuperfamily: tRNA-guanine transglycosylaseFamily: tRNA-guanine transglycosylase19 d1jm7a_
37.1
21
Fold: RING/U-boxSuperfamily: RING/U-boxFamily: RING finger domain, C3HC420 d1nexa1
35.8
21
Fold: Skp1 dimerisation domain-likeSuperfamily: Skp1 dimerisation domain-likeFamily: Skp1 dimerisation domain-like21 c2ashB_
not modelled
35.5
25
PDB header: transferaseChain: B: PDB Molecule: queuine trna-ribosyltransferase;PDBTitle: crystal structure of queuine trna-ribosyltransferase (ec 2.4.2.29)2 (trna-guanine (tm1561) from thermotoga maritima at 1.90 a resolution
22 d1ef4a_
not modelled
35.4
21
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: RNA polymerase subunit RPB10Family: RNA polymerase subunit RPB1023 c2jq5A_
not modelled
35.1
14
PDB header: structural genomicsChain: A: PDB Molecule: sec-c motif;PDBTitle: solution structure of rpa3114, a sec-c motif containing2 protein from rhodopseudomonas palustris; northeast3 structural genomics consortium target rpt5 / ontario4 center for structural proteomics target rp3097
24 d6paxa1
not modelled
33.7
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Paired domain25 c2eyqA_
not modelled
32.9
33
PDB header: hydrolaseChain: A: PDB Molecule: transcription-repair coupling factor;PDBTitle: crystal structure of escherichia coli transcription-repair2 coupling factor
26 c1u78A_
not modelled
30.8
14
PDB header: dna binding protein/dnaChain: A: PDB Molecule: transposable element tc3 transposase;PDBTitle: structure of the bipartite dna-binding domain of tc32 transposase bound to transposon dna
27 c3floD_
not modelled
29.1
21
PDB header: transferaseChain: D: PDB Molecule: dna polymerase alpha catalytic subunit a;PDBTitle: crystal structure of the carboxyl-terminal domain of yeast2 dna polymerase alpha in complex with its b subunit
28 d2gmga1
not modelled
28.6
42
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: PF0610-like29 d1wgna_
not modelled
27.0
22
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain30 c2opfA_
not modelled
26.3
67
PDB header: hydrolase/dnaChain: A: PDB Molecule: endonuclease viii;PDBTitle: crystal structure of the dna repair enzyme endonuclease-viii (nei)2 from e. coli (r252a) in complex with ap-site containing dna substrate
31 c3b40A_
not modelled
25.1
17
PDB header: hydrolaseChain: A: PDB Molecule: probable dipeptidase;PDBTitle: crystal structure of the probable dipeptidase pvdm from2 pseudomonas aeruginosa
32 d1k78a1
not modelled
24.4
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Paired domain33 d1p7ia_
not modelled
23.9
25
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain34 c3hosA_
not modelled
23.3
21
PDB header: transferase, dna binding protein/dnaChain: A: PDB Molecule: transposable element mariner, complete cds;PDBTitle: crystal structure of the mariner mos1 paired end complex with mg
35 c2qeuA_
not modelled
22.7
8
PDB header: lyaseChain: A: PDB Molecule: putative carboxymuconolactone decarboxylase;PDBTitle: crystal structure of putative carboxymuconolactone decarboxylase2 (yp_555818.1) from burkholderia xenovorans lb400 at 1.65 a resolution
36 c3llkA_
not modelled
22.7
28
PDB header: oxidoreductaseChain: A: PDB Molecule: sulfhydryl oxidase 1;PDBTitle: sulfhydryl oxidase fragment of human qsox1
37 c3iz6X_
not modelled
22.7
24
PDB header: ribosomeChain: X: PDB Molecule: 40s ribosomal protein s27 (s27e);PDBTitle: localization of the small subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
38 c3g2bA_
not modelled
21.9
8
PDB header: biosynthetic proteinChain: A: PDB Molecule: coenzyme pqq synthesis protein d;PDBTitle: crystal structure of pqqd from xanthomonas campestris
39 d1od6a_
not modelled
21.0
12
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Nucleotidylyl transferaseFamily: Adenylyltransferase40 d2ovra1
not modelled
20.9
32
Fold: Skp1 dimerisation domain-likeSuperfamily: Skp1 dimerisation domain-likeFamily: Skp1 dimerisation domain-like41 d1qxfa_
not modelled
20.5
18
Fold: Rubredoxin-likeSuperfamily: Zn-binding ribosomal proteinsFamily: Ribosomal protein S27e42 c2ragB_
not modelled
20.0
16
PDB header: hydrolaseChain: B: PDB Molecule: dipeptidase;PDBTitle: crystal structure of aminohydrolase from caulobacter crescentus
43 c3b9bA_
not modelled
20.0
12
PDB header: hydrolaseChain: A: PDB Molecule: sarcoplasmic/endoplasmic reticulum calciumPDBTitle: structure of the e2 beryllium fluoride complex of the serca2 ca2+-atpase
44 d1eb7a2
not modelled
20.0
22
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Di-heme cytochrome c peroxidase45 c1ug3A_
not modelled
19.8
13
PDB header: translationChain: A: PDB Molecule: eukaryotic protein synthesis initiation factorPDBTitle: c-terminal portion of human eif4gi
46 c2jnhA_
not modelled
19.7
13
PDB header: ligaseChain: A: PDB Molecule: e3 ubiquitin-protein ligase cbl-b;PDBTitle: solution structure of the uba domain from cbl-b
47 d1p91a_
not modelled
19.4
31
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: rRNA methyltransferase RlmA48 d1odfa_
not modelled
19.3
26
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Phosphoribulokinase/pantothenate kinase49 d2ga1a1
not modelled
18.9
10
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Alr1493-like50 c1nexC_
not modelled
18.7
21
PDB header: ligase, cell cycleChain: C: PDB Molecule: centromere dna-binding protein complex cbf3PDBTitle: crystal structure of scskp1-sccdc4-cpd peptide complex
51 c1ztbA_
not modelled
18.7
15
PDB header: ligaseChain: A: PDB Molecule: chorismate synthase;PDBTitle: crystal structure of chorismate synthase from mycobacterium2 tuberculosis
52 c3o0rC_
not modelled
18.6
13
PDB header: immune system/oxidoreductaseChain: C: PDB Molecule: nitric oxide reductase subunit c;PDBTitle: crystal structure of nitric oxide reductase from pseudomonas2 aeruginosa in complex with antibody fragment
53 c2pmzN_
not modelled
18.2
18
PDB header: translation, transferaseChain: N: PDB Molecule: dna-directed rna polymerase subunit n;PDBTitle: archaeal rna polymerase from sulfolobus solfataricus
54 c2jr6A_
not modelled
18.0
33
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0434 protein nma0874;PDBTitle: solution structure of upf0434 protein nma0874. northeast structural2 genomics target mr32
55 c3lrqB_
not modelled
18.0
30
PDB header: ligaseChain: B: PDB Molecule: e3 ubiquitin-protein ligase trim37;PDBTitle: crystal structure of the u-box domain of human ubiquitin-2 protein ligase (e3), northeast structural genomics3 consortium target hr4604d.
56 c2js4A_
not modelled
17.9
44
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0434 protein bb2007;PDBTitle: solution nmr structure of bordetella bronchiseptica protein2 bb2007. northeast structural genomics consortium target3 bor54
57 c2jvwA_
not modelled
17.4
25
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of uncharacterized protein q5e7h1 from vibrio2 fischeri. northeast structural genomics target vfr117
58 c2xzmO_
not modelled
17.2
10
PDB header: ribosomeChain: O: PDB Molecule: rps13e;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
59 d1y0ua_
not modelled
17.1
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: ArsR-like transcriptional regulators60 d2cwqa1
not modelled
17.0
13
Fold: AhpD-likeSuperfamily: AhpD-likeFamily: TTHA0727-like61 c3axjB_
not modelled
16.9
7
PDB header: dna binding proteinChain: B: PDB Molecule: translin associated factor x, isoform b;PDBTitle: high resolution crystal structure of c3po
62 c2p1nD_
not modelled
16.3
32
PDB header: signaling proteinChain: D: PDB Molecule: skp1-like protein 1a;PDBTitle: mechanism of auxin perception by the tir1 ubiqutin ligase
63 d1lv3a_
not modelled
16.1
22
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Hypothetical zinc finger protein YacG64 c2ecgA_
not modelled
16.0
9
PDB header: apoptosisChain: A: PDB Molecule: baculoviral iap repeat-containing protein 4;PDBTitle: solution structure of the ring domain of the baculoviral2 iap repeat-containing protein 4 from homo sapiens
65 d2lfba_
not modelled
16.0
21
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain66 d1sxjd1
not modelled
15.3
14
Fold: post-AAA+ oligomerization domain-likeSuperfamily: post-AAA+ oligomerization domain-likeFamily: DNA polymerase III clamp loader subunits, C-terminal domain67 c3itcA_
not modelled
15.2
4
PDB header: hydrolaseChain: A: PDB Molecule: renal dipeptidase;PDBTitle: crystal structure of sco3058 with bound citrate and glycerol
68 d2gmya1
not modelled
15.2
14
Fold: AhpD-likeSuperfamily: AhpD-likeFamily: Atu0492-like69 d2c0sa1
not modelled
14.9
43
Fold: ROP-likeSuperfamily: BAS1536-likeFamily: BAS1536-like70 c3s93B_
not modelled
14.5
12
PDB header: transcriptionChain: B: PDB Molecule: tudor domain-containing protein 5;PDBTitle: crystal structure of conserved motif in tdrd5
71 d1itua_
not modelled
14.5
12
Fold: TIM beta/alpha-barrelSuperfamily: Metallo-dependent hydrolasesFamily: Renal dipeptidase72 c3a44D_
not modelled
14.5
36
PDB header: metal binding proteinChain: D: PDB Molecule: hydrogenase nickel incorporation protein hypa;PDBTitle: crystal structure of hypa in the dimeric form
73 c2d9sA_
not modelled
14.4
10
PDB header: ligaseChain: A: PDB Molecule: cbl e3 ubiquitin protein ligase;PDBTitle: solution structure of rsgi ruh-049, a uba domain from mouse2 cdna
74 d1dl6a_
not modelled
14.4
25
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: Transcriptional factor domain75 c3izbX_
not modelled
14.3
33
PDB header: ribosomeChain: X: PDB Molecule: 40s ribosomal protein rps27 (s27e);PDBTitle: localization of the small subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
76 c3lpeF_
not modelled
14.3
24
PDB header: transferaseChain: F: PDB Molecule: dna-directed rna polymerase subunit e'';PDBTitle: crystal structure of spt4/5ngn heterodimer complex from methanococcus2 jannaschii
77 c1b6aA_
not modelled
14.3
21
PDB header: angiogenesis inhibitorChain: A: PDB Molecule: methionine aminopeptidase;PDBTitle: human methionine aminopeptidase 2 complexed with tnp-470
78 d2c2vv1
not modelled
14.2
13
Fold: RING/U-boxSuperfamily: RING/U-boxFamily: U-box79 c3pjaK_
not modelled
13.9
13
PDB header: hydrolaseChain: K: PDB Molecule: translin-associated protein x;PDBTitle: crystal structure of human c3po complex
80 d2cqxa1
not modelled
13.8
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain81 c3qw4B_
not modelled
13.8
15
PDB header: transferase, lyaseChain: B: PDB Molecule: ump synthase;PDBTitle: structure of leishmania donovani ump synthase
82 c1dl0A_
not modelled
13.7
46
PDB header: toxinChain: A: PDB Molecule: j-atracotoxin-hv1c;PDBTitle: solution structure of the insecticidal neurotoxin j-2 atracotoxin-hv1c
83 d1dl0a_
not modelled
13.7
46
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: omega toxin-likeFamily: Spider toxins84 d2jnya1
not modelled
13.6
33
Fold: Trm112p-likeSuperfamily: Trm112p-likeFamily: Trm112p-like85 c2xzm6_
not modelled
13.4
47
PDB header: ribosomeChain: 6: PDB Molecule: rps27e;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
86 c1nl0G_
not modelled
13.3
50
PDB header: immune systemChain: G: PDB Molecule: factor ix;PDBTitle: crystal structure of human factor ix gla domain in complex2 of an inhibitory antibody, 10c12
87 d1jgga_
not modelled
13.1
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain88 c3l6aA_
not modelled
13.0
5
PDB header: translationChain: A: PDB Molecule: eukaryotic translation initiation factor 4 gamma 2;PDBTitle: crystal structure of the c-terminal region of human p97
89 d1o6ba_
not modelled
12.9
8
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Nucleotidylyl transferaseFamily: Adenylyltransferase90 c3fdgA_
not modelled
12.6
16
PDB header: hydrolaseChain: A: PDB Molecule: dipeptidase ac. metallo peptidase. merops family m19;PDBTitle: the crystal structure of the dipeptidase ac, metallo peptidase. merops2 family m19
91 d1xkpa1
not modelled
12.3
14
Fold: Type III secretion system domainSuperfamily: Type III secretion system domainFamily: LcrE-like92 c2ao9H_
not modelled
12.2
13
PDB header: structural genomics, unknown functionChain: H: PDB Molecule: phage protein;PDBTitle: structural genomics, the crystal structure of a phage protein2 (phbc6a51) from bacillus cereus atcc 14579
93 c2kpiA_
not modelled
12.1
44
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein sco3027;PDBTitle: solution nmr structure of streptomyces coelicolor sco30272 modeled with zn+2 bound, northeast structural genomics3 consortium target rr58
94 d2ao9a1
not modelled
12.0
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Nanomeric phage protein-like95 c1ee8A_
not modelled
11.9
57
PDB header: dna binding proteinChain: A: PDB Molecule: mutm (fpg) protein;PDBTitle: crystal structure of mutm (fpg) protein from thermus thermophilus hb8
96 d1d8ja_
not modelled
11.9
33
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: The central core domain of TFIIE beta97 c3bg3B_
not modelled
11.9
10
PDB header: ligaseChain: B: PDB Molecule: pyruvate carboxylase, mitochondrial;PDBTitle: crystal structure of human pyruvate carboxylase (missing2 the biotin carboxylase domain at the n-terminus)
98 c2do6A_
not modelled
11.9
15
PDB header: ligaseChain: A: PDB Molecule: e3 ubiquitin-protein ligase cbl-b;PDBTitle: solution structure of rsgi ruh-065, a uba domain from human2 cdna
99 d1vnda_
not modelled
11.8
8
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain