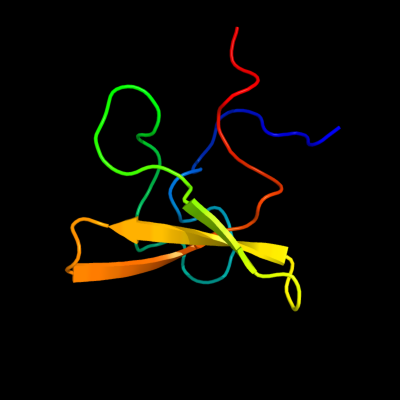
1 c2kt8A_
99.0
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: probable surface protein;PDBTitle: solution nmr structure of the cpe1231(468-535) protein from2 clostridium perfringens, northeast structural genomics3 consortium target cpr82b
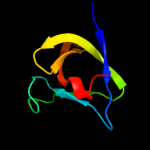
2 c2krsA_
98.9
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: probable enterotoxin;PDBTitle: solution nmr structure of sh3 domain from cpf_0587 (fragment2 415-479) from clostridium perfringens. northeast structural3 genomics consortium (nesg) target cpr74a.
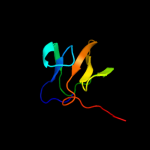
3 c2kybA_
98.6
15
PDB header: toxinChain: A: PDB Molecule: mannosyl-glycoprotein endo-beta-n-acetylglucosamidasePDBTitle: solution structure of cpr82g from clostridium perfringens. north east2 structural genomics consortium target cpr82g
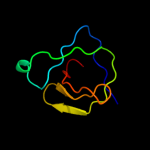
4 c2kq8A_
98.6
28
PDB header: hydrolaseChain: A: PDB Molecule: cell wall hydrolase;PDBTitle: solution nmr structure of a domain from bt9727_4915 from2 bacillus thuringiensis, northeast structural genomics3 consortium target bur95a
5 c3npfB_
98.4
16
PDB header: hydrolaseChain: B: PDB Molecule: putative dipeptidyl-peptidase vi;PDBTitle: crystal structure of a putative dipeptidyl-peptidase vi (bacova_00612)2 from bacteroides ovatus at 1.72 a resolution
6 c3h41A_
97.4
24
PDB header: hydrolaseChain: A: PDB Molecule: nlp/p60 family protein;PDBTitle: crystal structure of a nlpc/p60 family protein (bce_2878) from2 bacillus cereus atcc 10987 at 1.79 a resolution
7 c1xovA_
96.5
10
PDB header: hydrolaseChain: A: PDB Molecule: ply protein;PDBTitle: the crystal structure of the listeria monocytogenes bacteriophage psa2 endolysin plypsa
8 c3m1uB_
96.5
18
PDB header: hydrolaseChain: B: PDB Molecule: putative gamma-d-glutamyl-l-diamino acid endopeptidase;PDBTitle: crystal structure of a putative gamma-d-glutamyl-l-diamino acid2 endopeptidase (dvu_0896) from desulfovibrio vulgaris hildenborough at3 1.75 a resolution
9 d1wfwa_
96.3
30
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain10 d1cska_
96.0
20
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain11 c3pe0B_
95.9
26
PDB header: structural proteinChain: B: PDB Molecule: plectin;PDBTitle: structure of the central region of the plakin domain of plectin
12 c2dx1A_
95.8
9
PDB header: signaling proteinChain: A: PDB Molecule: rho guanine nucleotide exchange factor 4;PDBTitle: crystal structure of rhogef protein asef
13 c3qwyA_
95.7
18
PDB header: signaling proteinChain: A: PDB Molecule: cell death abnormality protein 2;PDBTitle: ced-2
14 c1k9aB_
95.7
20
PDB header: transferaseChain: B: PDB Molecule: carboxyl-terminal src kinase;PDBTitle: crystal structure analysis of full-length carboxyl-terminal2 src kinase at 2.5 a resolution
15 d1gl5a_
95.7
20
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain16 c3qwxX_
95.6
18
PDB header: signaling proteinChain: X: PDB Molecule: cell death abnormality protein 2;PDBTitle: ced-2 1-174
17 c2x3wD_
95.6
21
PDB header: endocytosisChain: D: PDB Molecule: protein kinase c and casein kinase substrate in neuronsPDBTitle: structure of mouse syndapin i (crystal form 2)
18 d1k9aa1
95.6
18
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain19 d1ckaa_
95.5
16
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain20 c2eyyA_
95.5
17
PDB header: signaling proteinChain: A: PDB Molecule: v-crk sarcoma virus ct10 oncogene homologPDBTitle: ct10-regulated kinase isoform i
21 c2ct4A_
not modelled
95.4
20
PDB header: signaling proteinChain: A: PDB Molecule: cdc42-interacting protein 4;PDBTitle: solution strutcure of the sh3 domain of the cdc42-2 interacting protein 4
22 d1ng2a2
not modelled
95.4
16
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain23 d1k1za_
not modelled
95.3
21
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain24 c1s1nA_
not modelled
95.2
21
PDB header: cell adhesionChain: A: PDB Molecule: nephrocystin 1;PDBTitle: sh3 domain of human nephrocystin
25 c1griA_
not modelled
95.1
14
PDB header: signal transduction adaptorChain: A: PDB Molecule: growth factor bound protein 2;PDBTitle: grb2
26 d1qlya_
not modelled
95.1
21
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain27 c2kbtA_
not modelled
95.1
17
PDB header: signaling proteinChain: A: PDB Molecule: chimera of proto-oncogene vav, linker,PDBTitle: attachment of an nmr-invisible solubility enhancement tag2 (inset) using a sortase-mediated protein ligation method
28 c2rf0D_
not modelled
95.1
23
PDB header: transferaseChain: D: PDB Molecule: mitogen-activated protein kinase kinase kinase 10;PDBTitle: crystal structure of human mixed lineage kinase map3k10 sh3 domain
29 c2dl5A_
not modelled
95.1
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: kiaa0769 protein;PDBTitle: solution structure of the first sh3 domain of human2 kiaa0769 protein
30 c1zlmA_
not modelled
95.1
24
PDB header: signaling proteinChain: A: PDB Molecule: osteoclast stimulating factor 1;PDBTitle: crystal structure of the sh3 domain of human osteoclast2 stimulating factor
31 d1opka1
not modelled
95.1
16
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain32 d1oeba_
not modelled
95.0
16
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain33 c2egcA_
not modelled
95.0
15
PDB header: signaling proteinChain: A: PDB Molecule: sh3 and px domain-containing protein 2a;PDBTitle: solution structure of the fifth sh3 domain from human2 kiaa0418 protein
34 c1z9zA_
not modelled
95.0
18
PDB header: structural proteinChain: A: PDB Molecule: cytoskeleton assembly control protein sla1;PDBTitle: crystal structure of yeast sla1 sh3 domain 3
35 c1ov3A_
not modelled
95.0
16
PDB header: oxidoreductase activatorChain: A: PDB Molecule: neutrophil cytosol factor 1;PDBTitle: structure of the p22phox-p47phox complex
36 d1nega_
not modelled
95.0
22
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain37 c1negA_
not modelled
95.0
22
PDB header: structural proteinChain: A: PDB Molecule: spectrin alpha chain, brain;PDBTitle: crystal structure analysis of n-and c-terminal labeled sh3-2 domain of alpha-chicken spectrin
38 d1uj0a_
not modelled
94.9
26
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain39 d1pwta_
not modelled
94.9
24
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain40 d1wlpb2
not modelled
94.9
16
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain41 c2xmfA_
not modelled
94.9
14
PDB header: motor proteinChain: A: PDB Molecule: myosin 1e sh3;PDBTitle: myosin 1e sh3
42 c1ei3E_
94.8
14
PDB header: PDB COMPND: 43 c1m9sA_
not modelled
94.8
13
PDB header: signaling proteinChain: A: PDB Molecule: internalin b;PDBTitle: crystal structure of internalin b (inlb), a listeria2 monocytogenes virulence protein containing sh3-like3 domains.
44 d1utia_
not modelled
94.8
16
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain45 c2azsA_
not modelled
94.8
12
PDB header: signaling proteinChain: A: PDB Molecule: sh2-sh3 adapter protein drk;PDBTitle: nmr structure of the n-terminal sh3 domain of drk2 (calculated without noe restraints)
46 c2fg0B_
not modelled
94.8
13
PDB header: hydrolaseChain: B: PDB Molecule: cog0791: cell wall-associated hydrolases (invasion-PDBTitle: crystal structure of a putative gamma-d-glutamyl-l-diamino acid2 endopeptidase (npun_r0659) from nostoc punctiforme pcc 73102 at 1.793 a resolution
47 c1x6gA_
not modelled
94.8
25
PDB header: signaling proteinChain: A: PDB Molecule: megakaryocyte-associated tyrosine-protein kinase;PDBTitle: solution structures of the sh3 domain of human2 megakaryocyte-associated tyrosine-protein kinase.
48 c2df6A_
not modelled
94.8
16
PDB header: signaling proteinChain: A: PDB Molecule: rho guanine nucleotide exchange factor 7;PDBTitle: crystal structure of the sh3 domain of betapix in complex2 with a high affinity peptide from pak2
49 c2j6kE_
not modelled
94.8
16
PDB header: protein bindingChain: E: PDB Molecule: cd2-associated protein;PDBTitle: n-terminal sh3 domain of cms (cd2ap human homolog)
50 d1fmka1
not modelled
94.8
18
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain51 d1u06a1
not modelled
94.8
24
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain52 c2ekhA_
not modelled
94.8
23
PDB header: signaling proteinChain: A: PDB Molecule: sh3 and px domain-containing protein 2a;PDBTitle: solution structures of the sh3 domain of human kiaa0418
53 c2epdA_
not modelled
94.8
18
PDB header: protein bindingChain: A: PDB Molecule: rho gtpase-activating protein 4;PDBTitle: solution structure of sh3 domain in rho-gtpase-activating2 protein 4
54 d1ov3a2
not modelled
94.8
16
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain55 c1zx6A_
not modelled
94.8
22
PDB header: protein bindingChain: A: PDB Molecule: ypr154wp;PDBTitle: high-resolution crystal structure of yeast pin3 sh3 domain
56 c2fo0A_
not modelled
94.7
18
PDB header: transferaseChain: A: PDB Molecule: proto-oncogene tyrosine-protein kinase abl1 (1b isoform);PDBTitle: organization of the sh3-sh2 unit in active and inactive forms of the2 c-abl tyrosine kinase
57 c1x2pA_
not modelled
94.7
17
PDB header: transferaseChain: A: PDB Molecule: protein arginine n-methyltransferase 2;PDBTitle: solution structure of the sh3 domain of the protein2 arginine n-methyltransferase 2
58 c2cucA_
not modelled
94.7
18
PDB header: signaling proteinChain: A: PDB Molecule: sh3 domain containing ring finger 2;PDBTitle: solution structure of the sh3 domain of the mouse2 hypothetical protein sh3rf2
59 c2enmA_
not modelled
94.7
21
PDB header: endocytosisChain: A: PDB Molecule: sorting nexin-9;PDBTitle: solution structure of the sh3 domain from mouse sorting2 nexin-9
60 c3cqtA_
not modelled
94.7
21
PDB header: transferaseChain: A: PDB Molecule: proto-oncogene tyrosine-protein kinase fyn;PDBTitle: n53i v55l mutant of fyn sh3 domain
61 c2dmoA_
not modelled
94.7
28
PDB header: signaling proteinChain: A: PDB Molecule: neutrophil cytosol factor 2;PDBTitle: refined solution structure of the 1st sh3 domain from human2 neutrophil cytosol factor 2 (ncf-2)
62 d1oota_
not modelled
94.6
19
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain63 d1gcqc_
not modelled
94.6
21
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain64 c1w70A_
not modelled
94.6
25
PDB header: sh3 domainChain: A: PDB Molecule: neutrophil cytosol factor 4;PDBTitle: sh3 domain of p40phox complexed with c-terminal polyproline2 region of p47phox
65 d1uuea_
not modelled
94.6
24
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain66 c1deqO_
94.6
13
PDB header: PDB COMPND: 67 d1k4us_
not modelled
94.6
15
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain68 c3ojaB_
94.6
11
PDB header: protein bindingChain: B: PDB Molecule: anopheles plasmodium-responsive leucine-rich repeat proteinPDBTitle: crystal structure of lrim1/apl1c complex
69 d1gria2
not modelled
94.6
14
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain70 c2ed0A_
not modelled
94.5
10
PDB header: signaling proteinChain: A: PDB Molecule: abl interactor 2;PDBTitle: solution structure of the sh3 domain of abl interactor 22 (abelson interactor 2)
71 d2rn8a1
not modelled
94.5
24
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain72 d2fo0a1
not modelled
94.5
16
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain73 d1e6ga_
not modelled
94.5
30
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain74 d1u5sa1
not modelled
94.5
14
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain75 c1oplA_
not modelled
94.5
18
PDB header: transferaseChain: A: PDB Molecule: proto-oncogene tyrosine-protein kinase;PDBTitle: structural basis for the auto-inhibition of c-abl tyrosine2 kinase
76 c2jxbA_
not modelled
94.5
21
PDB header: signaling protein complexChain: A: PDB Molecule: t-cell surface glycoprotein cd3 epsilon chain,PDBTitle: structure of cd3epsilon-nck2 first sh3 domain complex
77 c2csqA_
not modelled
94.5
11
PDB header: endocytosis/exocytosisChain: A: PDB Molecule: rim binding protein 2;PDBTitle: solution structure of the second sh3 domain of human rim-2 binding protein 2
78 c2d1xD_
not modelled
94.5
11
PDB header: cell invasionChain: D: PDB Molecule: cortactin isoform a;PDBTitle: the crystal structure of the cortactin-sh3 domain and amap1-2 peptide complex
79 d1tuca_
not modelled
94.5
24
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain80 d1sema_
not modelled
94.5
19
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain81 c1ng2A_
not modelled
94.4
15
PDB header: oxidoreductase activatorChain: A: PDB Molecule: neutrophil cytosolic factor 1;PDBTitle: structure of autoinhibited p47phox
82 c1zuyB_
not modelled
94.4
8
PDB header: contractile proteinChain: B: PDB Molecule: myosin-5 isoform;PDBTitle: high-resolution structure of yeast myo5 sh3 domain
83 c2nwmA_
not modelled
94.4
20
PDB header: cell adhesionChain: A: PDB Molecule: vinexin;PDBTitle: solution structure of the first sh3 domain of human vinexin2 and its interaction with the peptides from vinculin
84 d1gcqa_
not modelled
94.3
14
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain85 d1e6ha_
not modelled
94.3
24
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain86 c2ablA_
not modelled
94.3
18
PDB header: transferaseChain: A: PDB Molecule: abl tyrosine kinase;PDBTitle: sh3-sh2 domain fragment of human bcr-abl tyrosine kinase
87 c1wxtA_
not modelled
94.3
32
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein flj21522;PDBTitle: solution structure of the sh3 domain of human hypothetical2 protein flj21522
88 c2l0aA_
not modelled
94.3
21
PDB header: signaling proteinChain: A: PDB Molecule: signal transducing adapter molecule 1;PDBTitle: solution nmr structure of signal transducing adapter molecule 1 stam-12 from homo sapiens, northeast structural genomics consortium target3 hr4479e
89 c2pqhA_
not modelled
94.3
23
PDB header: structural proteinChain: A: PDB Molecule: spectrin alpha chain, brain;PDBTitle: structure of sh3 chimera with a type ii ligand linked to the chain c-2 terminal
90 c2js0A_
not modelled
94.3
15
PDB header: signaling proteinChain: A: PDB Molecule: cytoplasmic protein nck1;PDBTitle: solution structure of second sh3 domain of adaptor nck
91 d1shfa_
not modelled
94.3
18
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain92 c2bz8B_
not modelled
94.2
19
PDB header: sh3 domainChain: B: PDB Molecule: sh3-domain kinase binding protein 1;PDBTitle: n-terminal sh3 domain of cin85 bound to cbl-b peptide
93 d1ycsb2
not modelled
94.2
13
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain94 c2k2mA_
not modelled
94.2
23
PDB header: signaling proteinChain: A: PDB Molecule: eps8-like protein 1;PDBTitle: structural basis of pxxdy motif recognition in sh3 binding
95 d1jo8a_
not modelled
94.2
11
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain96 c1y57A_
not modelled
94.2
18
PDB header: transferaseChain: A: PDB Molecule: proto-oncogene tyrosine-protein kinase src;PDBTitle: structure of unphosphorylated c-src in complex with an inhibitor
97 c2h8hA_
not modelled
94.2
18
PDB header: transferaseChain: A: PDB Molecule: proto-oncogene tyrosine-protein kinase src;PDBTitle: src kinase in complex with a quinazoline inhibitor
98 c2ct3A_
not modelled
94.2
11
PDB header: signaling proteinChain: A: PDB Molecule: vinexin;PDBTitle: solution structure of the sh3 domain of the vinexin protein
99 c2egaA_
not modelled
94.1
24
PDB header: signaling proteinChain: A: PDB Molecule: sh3 and px domain-containing protein 2a;PDBTitle: solution structure of the first sh3 domain from human2 kiaa0418 protein
100 d1h8ka_
not modelled
94.1
32
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain101 c2c0iA_
not modelled
94.1
14
PDB header: transferaseChain: A: PDB Molecule: tyrosine-protein kinase hck;PDBTitle: src family kinase hck with bound inhibitor a-420983
102 c2yuoA_
not modelled
94.0
13
PDB header: signaling proteinChain: A: PDB Molecule: run and tbc1 domain containing 3;PDBTitle: solution structure of the sh3 domain of mouse run and tbc12 domain containing 3
103 c2yunA_
not modelled
94.0
15
PDB header: protein transportChain: A: PDB Molecule: nostrin;PDBTitle: solution structure of the sh3 domain of human nostrin
104 c2drmB_
not modelled
94.0
19
PDB header: contractile proteinChain: B: PDB Molecule: acanthamoeba myosin ib;PDBTitle: acanthamoeba myosin i sh3 domain bound to acan125
105 c1wxbA_
not modelled
94.0
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: epidermal growth factor receptor pathwayPDBTitle: solution structure of the sh3 domain from human epidermal2 growth factor receptor pathway substrate 8-like protein
106 c1u3oA_
not modelled
94.0
21
PDB header: signaling proteinChain: A: PDB Molecule: huntingtin-associated protein-interactingPDBTitle: solution structure of rat kalirin n-terminal sh3 domain
107 c2dnuA_
not modelled
94.0
14
PDB header: structural genomics, structural proteinChain: A: PDB Molecule: sh3 multiple domains 1;PDBTitle: solution structure of rsgi ruh-061, a sh3 domain from human
108 c2jw4A_
not modelled
93.9
23
PDB header: signaling proteinChain: A: PDB Molecule: cytoplasmic protein nck1;PDBTitle: nmr solution structure of the n-terminal sh3 domain of2 human nckalpha
109 d2v1ra1
not modelled
93.9
16
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain110 c2dilA_
not modelled
93.9
16
PDB header: cell adhesionChain: A: PDB Molecule: proline-serine-threonine phosphatase-interactingPDBTitle: solution structure of the sh3 domain of the human proline-2 serine-threonine phosphatase-interacting protein 1
111 d1lcka1
not modelled
93.9
17
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain112 c2a28D_
not modelled
93.7
14
PDB header: signaling proteinChain: D: PDB Molecule: bzz1 protein;PDBTitle: atomic-resolution crystal structure of the second sh32 domain of yeast bzz1 determined from a pseudomerohedrally3 twinned crystal
113 c1x27F_
not modelled
93.6
18
PDB header: signaling proteinChain: F: PDB Molecule: proto-oncogene tyrosine-protein kinase lck;PDBTitle: crystal structure of lck sh2-sh3 with sh2 binding site of2 p130cas
114 c1x2qA_
not modelled
93.6
14
PDB header: signaling proteinChain: A: PDB Molecule: signal transducing adapter molecule 2;PDBTitle: solution structure of the sh3 domain of the signal2 transducing adaptor molecule 2
115 d1gbra_
not modelled
93.6
17
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain116 c2wpqA_
93.6
8
PDB header: membrane proteinChain: A: PDB Molecule: trimeric autotransporter adhesin fragment;PDBTitle: salmonella enterica sada 479-519 fused to gcn4 adaptors (2 sadak3, in-register fusion)
117 c2djqA_
not modelled
93.6
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: sh3 domain containing ring finger 2;PDBTitle: the solution structure of the first sh3 domain of mouse sh32 domain containing ring finger 2
118 d1ov3a1
not modelled
93.6
16
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain119 c1deqF_
93.6
6
PDB header: PDB COMPND: 120 c2eqiA_
not modelled
93.6
8
PDB header: immune system, hydrolaseChain: A: PDB Molecule: phospholipase c, gamma 2;PDBTitle: solution structure of the sh3 domain from phospholipase c,2 gamma 2