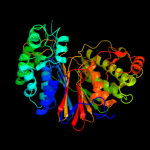
1 d2nlza1
100.0
31
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Gamma-glutamyltranspeptidase-like2 c2e0wA_
100.0
100
PDB header: transferaseChain: A: PDB Molecule: gamma-glutamyltranspeptidase;PDBTitle: t391a precursor mutant protein of gamma-glutamyltranspeptidase from2 escherichia coli
3 d2i3oa1
100.0
24
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Gamma-glutamyltranspeptidase-like4 c2z8jA_
100.0
100
PDB header: transferaseChain: A: PDB Molecule: gamma-glutamyltranspeptidase;PDBTitle: crystal structure of escherichia coli gamma-2 glutamyltranspeptidase in complex with azaserine prepared3 in the dark
5 c2v36A_
100.0
40
PDB header: transferaseChain: A: PDB Molecule: gamma-glutamyltranspeptidase large chain;PDBTitle: crystal structure of gamma-glutamyl transferase from2 bacillus subtilis
6 c2qm6C_
100.0
49
PDB header: transferaseChain: C: PDB Molecule: gamma-glutamyltranspeptidase;PDBTitle: crystal structure of helicobacter pylori gamma-glutamyltranspeptidase2 in complex with glutamate
7 c3g9kD_
100.0
38
PDB header: hydrolaseChain: D: PDB Molecule: capsule biosynthesis protein capd;PDBTitle: crystal structure of bacillus anthracis transpeptidase enzyme capd
8 c2e0yB_
100.0
96
PDB header: transferaseChain: B: PDB Molecule: gamma-glutamyltranspeptidase;PDBTitle: crystal structure of the samarium derivative of mature gamma-2 glutamyltranspeptidase from escherichia coli
9 c2nqoB_
100.0
58
PDB header: transferaseChain: B: PDB Molecule: gamma-glutamyltranspeptidase;PDBTitle: crystal structure of helicobacter pylori gamma-glutamyltranspeptidase
10 c2v36D_
100.0
42
PDB header: transferaseChain: D: PDB Molecule: gamma-glutamyltranspeptidase small chain;PDBTitle: crystal structure of gamma-glutamyl transferase from2 bacillus subtilis
11 c3ga9S_
100.0
33
PDB header: hydrolaseChain: S: PDB Molecule: capsule biosynthesis protein capd;PDBTitle: crystal structure of bacillus anthracis transpeptidase enzyme capd,2 crystal form ii
12 c1apyA_
94.9
25
PDB header: hydrolaseChain: A: PDB Molecule: aspartylglucosaminidase;PDBTitle: human aspartylglucosaminidase
13 c1t3mA_
94.9
18
PDB header: hydrolaseChain: A: PDB Molecule: putative l-asparaginase;PDBTitle: structure of the isoaspartyl peptidase with l-asparaginase2 activity from e. coli
14 c2gezE_
94.4
23
PDB header: hydrolaseChain: E: PDB Molecule: l-asparaginase alpha subunit;PDBTitle: crystal structure of potassium-independent plant asparaginase
15 c2zakB_
93.4
18
PDB header: hydrolaseChain: B: PDB Molecule: l-asparaginase precursor;PDBTitle: orthorhombic crystal structure of precursor e. coli isoaspartyl2 peptidase/l-asparaginase (ecaiii) with active-site t179a mutation
16 c1p4vA_
92.9
18
PDB header: hydrolaseChain: A: PDB Molecule: n(4)-(beta-n-acetylglucosaminyl)-l-asparaginasePDBTitle: crystal structure of the glycosylasparaginase precursor2 d151n mutant with glycine
17 c2gacA_
92.4
26
PDB header: hydrolaseChain: A: PDB Molecule: glycosylasparaginase;PDBTitle: t152c mutant glycosylasparaginase from flavobacterium2 meningosepticum
18 c2a8lB_
92.2
26
PDB header: hydrolaseChain: B: PDB Molecule: threonine aspartase 1;PDBTitle: crystal structure of human taspase1 (t234a mutant)
19 c1apzB_
85.9
19
PDB header: complex (hydrolase/peptide)Chain: B: PDB Molecule: aspartylglucosaminidase;PDBTitle: human aspartylglucosaminidase complex with reaction product
20 c3cuqA_
85.6
45
PDB header: protein transportChain: A: PDB Molecule: vacuolar-sorting protein snf8;PDBTitle: integrated structural and functional model of the human escrt-ii2 complex
21 c2zmeA_
not modelled
85.4
45
PDB header: protein transportChain: A: PDB Molecule: vacuolar-sorting protein snf8;PDBTitle: integrated structural and functional model of the human escrt-ii2 complex
22 c1u5tA_
not modelled
84.8
28
PDB header: transport proteinChain: A: PDB Molecule: appears to be functionally related to snf7;PDBTitle: structure of the escrt-ii endosomal trafficking complex
23 d1u5ta1
not modelled
78.9
26
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Vacuolar sorting protein domain24 c1w7pD_
not modelled
72.5
33
PDB header: protein transportChain: D: PDB Molecule: vps36p, ylr417w;PDBTitle: the crystal structure of endosomal complex escrt-ii2 (vps22/vps25/vps36)
25 c1u5tB_
not modelled
69.7
37
PDB header: transport proteinChain: B: PDB Molecule: defective in vacuolar protein sorting; vps36p;PDBTitle: structure of the escrt-ii endosomal trafficking complex
26 c2zmeB_
not modelled
69.2
19
PDB header: protein transportChain: B: PDB Molecule: vacuolar protein-sorting-associated protein 36;PDBTitle: integrated structural and functional model of the human escrt-ii2 complex
27 d1jq5a_
not modelled
64.7
23
Fold: Dehydroquinate synthase-likeSuperfamily: Dehydroquinate synthase-likeFamily: Iron-containing alcohol dehydrogenase28 c1t3mD_
not modelled
62.1
27
PDB header: hydrolaseChain: D: PDB Molecule: putative l-asparaginase;PDBTitle: structure of the isoaspartyl peptidase with l-asparaginase2 activity from e. coli
29 c1jn9B_
not modelled
62.1
27
PDB header: hydrolaseChain: B: PDB Molecule: putative l-asparaginase;PDBTitle: structure of putative asparaginase encoded by escherichia coli ybik2 gene
30 c1t3mB_
not modelled
62.1
27
PDB header: hydrolaseChain: B: PDB Molecule: putative l-asparaginase;PDBTitle: structure of the isoaspartyl peptidase with l-asparaginase2 activity from e. coli
31 c2zalD_
not modelled
58.9
27
PDB header: hydrolaseChain: D: PDB Molecule: l-asparaginase;PDBTitle: crystal structure of e. coli isoaspartyl aminopeptidase/l-asparaginase2 in complex with l-aspartate
32 c2gacD_
not modelled
55.0
19
PDB header: hydrolaseChain: D: PDB Molecule: glycosylasparaginase;PDBTitle: t152c mutant glycosylasparaginase from flavobacterium2 meningosepticum
33 c2gezF_
not modelled
53.6
21
PDB header: hydrolaseChain: F: PDB Molecule: l-asparaginase beta subunit;PDBTitle: crystal structure of potassium-independent plant asparaginase
34 c1k2xB_
not modelled
51.1
27
PDB header: hydrolaseChain: B: PDB Molecule: putative l-asparaginase;PDBTitle: crystal structure of putative asparaginase encoded by escherichia coli2 ybik gene
35 c1k2xD_
not modelled
51.1
27
PDB header: hydrolaseChain: D: PDB Molecule: putative l-asparaginase;PDBTitle: crystal structure of putative asparaginase encoded by escherichia coli2 ybik gene
36 c1jn9D_
not modelled
51.1
27
PDB header: hydrolaseChain: D: PDB Molecule: putative l-asparaginase;PDBTitle: structure of putative asparaginase encoded by escherichia coli ybik2 gene
37 c2zalB_
not modelled
48.3
27
PDB header: hydrolaseChain: B: PDB Molecule: l-asparaginase;PDBTitle: crystal structure of e. coli isoaspartyl aminopeptidase/l-asparaginase2 in complex with l-aspartate
38 c3rf7A_
not modelled
31.4
16
PDB header: oxidoreductaseChain: A: PDB Molecule: iron-containing alcohol dehydrogenase;PDBTitle: crystal structure of an iron-containing alcohol dehydrogenase2 (sden_2133) from shewanella denitrificans os-217 at 2.12 a resolution
39 c2wl8D_
not modelled
31.2
19
PDB header: protein transportChain: D: PDB Molecule: peroxisomal biogenesis factor 19;PDBTitle: x-ray crystal structure of pex19p
40 d2fi0a1
not modelled
28.9
19
Fold: SP0561-likeSuperfamily: SP0561-likeFamily: SP0561-like41 d1u2ka_
not modelled
26.3
11
Fold: Heme-dependent peroxidasesSuperfamily: Heme-dependent peroxidasesFamily: Catalase-peroxidase KatG42 c3bt3B_
not modelled
25.4
13
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: glyoxalase-related enzyme, arac type;PDBTitle: crystal structure of a glyoxalase-related enzyme from clostridium2 phytofermentans
43 c3h7hA_
not modelled
24.1
11
PDB header: transcriptionChain: A: PDB Molecule: transcription elongation factor spt4;PDBTitle: crystal structure of the human transcription elongation factor dsif,2 hspt4/hspt5 (176-273)
44 c3gxqB_
not modelled
23.9
50
PDB header: dna binding protein/dnaChain: B: PDB Molecule: putative regulator of transfer genes arta;PDBTitle: structure of arta and dna complex
45 d1o2da_
not modelled
23.7
27
Fold: Dehydroquinate synthase-likeSuperfamily: Dehydroquinate synthase-likeFamily: Iron-containing alcohol dehydrogenase46 d2ccaa1
not modelled
23.3
15
Fold: Heme-dependent peroxidasesSuperfamily: Heme-dependent peroxidasesFamily: Catalase-peroxidase KatG47 c1udsA_
not modelled
21.1
17
PDB header: transferaseChain: A: PDB Molecule: ribonuclease ph;PDBTitle: crystal structure of the trna processing enzyme rnase ph r126a mutant2 from aquifex aeolicus
48 d1oj7a_
not modelled
20.8
20
Fold: Dehydroquinate synthase-likeSuperfamily: Dehydroquinate synthase-likeFamily: Iron-containing alcohol dehydrogenase49 c2kmvA_
not modelled
20.5
13
PDB header: hydrolaseChain: A: PDB Molecule: copper-transporting atpase 1;PDBTitle: solution structure of the nucleotide binding domain of the2 human menkes protein in the atp-free form
50 c3hkmB_
not modelled
19.9
26
PDB header: hydrolaseChain: B: PDB Molecule: os03g0854200 protein;PDBTitle: crystal structure of rice(oryza sativa) rrp46
51 d1u5tb1
not modelled
19.4
33
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Vacuolar sorting protein domain52 c2qntA_
not modelled
19.0
25
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein atu1872;PDBTitle: crystal structure of protein of unknown function from agrobacterium2 tumefaciens str. c58
53 d2nn6f1
not modelled
17.6
33
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Ribonuclease PH domain 1-like54 d1r6la1
not modelled
17.2
11
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Ribonuclease PH domain 1-like55 c2nn6D_
not modelled
16.9
21
PDB header: hydrolase/transferaseChain: D: PDB Molecule: exosome complex exonuclease rrp46;PDBTitle: structure of the human rna exosome composed of rrp41, rrp45,2 rrp46, rrp43, mtr3, rrp42, csl4, rrp4, and rrp40
56 d2nn6b1
not modelled
16.6
22
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Ribonuclease PH domain 1-like57 c3b4tC_
not modelled
16.4
32
PDB header: transferaseChain: C: PDB Molecule: ribonuclease ph;PDBTitle: crystal structure of mycobacterium tuberculosis rnase ph, the2 mycobacterium tuberculosis structural genomics consortium target3 rv1340
58 d1udsa1
not modelled
16.1
17
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Ribonuclease PH domain 1-like59 d2nn6a1
not modelled
15.9
33
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Ribonuclease PH domain 1-like60 d1a0da_
not modelled
15.7
24
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase61 d2nn6e1
not modelled
15.6
39
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Ribonuclease PH domain 1-like62 d1vlja_
not modelled
15.6
25
Fold: Dehydroquinate synthase-likeSuperfamily: Dehydroquinate synthase-likeFamily: Iron-containing alcohol dehydrogenase63 d2je6b1
not modelled
15.6
11
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Ribonuclease PH domain 1-like64 c1u2jC_
not modelled
15.6
9
PDB header: oxidoreductaseChain: C: PDB Molecule: peroxidase/catalase hpi;PDBTitle: crystal structure of the c-terminal domain from the2 catalase-peroxidase katg of escherichia coli (p21 21 21)
65 c2rk9B_
not modelled
14.8
14
PDB header: oxidoreductaseChain: B: PDB Molecule: glyoxalase/bleomycin resistance protein/dioxygenase;PDBTitle: the crystal structure of a glyoxalase/bleomycin resistance2 protein/dioxygenase superfamily member from vibrio splendidus 12b01
66 d2ba0g1
not modelled
13.9
33
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Ribonuclease PH domain 1-like67 c3itwA_
not modelled
13.8
26
PDB header: peptide binding proteinChain: A: PDB Molecule: protein tiox;PDBTitle: crystal structure of tiox from micromonospora sp. ml1
68 d2nn6d1
not modelled
13.5
6
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Ribonuclease PH domain 1-like69 c2wnrB_
not modelled
13.3
32
PDB header: hydrolaseChain: B: PDB Molecule: probable exosome complex exonuclease 1;PDBTitle: the structure of methanothermobacter thermautotrophicus2 exosome core assembly
70 d1kq3a_
not modelled
12.4
20
Fold: Dehydroquinate synthase-likeSuperfamily: Dehydroquinate synthase-likeFamily: Iron-containing alcohol dehydrogenase71 c2nn6F_
not modelled
12.1
33
PDB header: hydrolase/transferaseChain: F: PDB Molecule: exosome component 6;PDBTitle: structure of the human rna exosome composed of rrp41, rrp45,2 rrp46, rrp43, mtr3, rrp42, csl4, rrp4, and rrp40
72 d1ny9a_
not modelled
12.1
11
Fold: Antibiotic binding domain of TipA-like multidrug resistance regulatorsSuperfamily: Antibiotic binding domain of TipA-like multidrug resistance regulatorsFamily: Antibiotic binding domain of TipA-like multidrug resistance regulators73 c1ny9A_
not modelled
12.1
11
PDB header: transcriptionChain: A: PDB Molecule: transcriptional activator tipa-s;PDBTitle: antibiotic binding domain of a tipa-class multidrug2 resistance transcriptional regulator
74 c2e5yA_
not modelled
11.9
17
PDB header: hydrolaseChain: A: PDB Molecule: atp synthase epsilon chain;PDBTitle: epsilon subunit and atp complex of f1f0-atp synthase from2 the thermophilic bacillus ps3
75 c2wp8B_
not modelled
11.9
11
PDB header: hydrolaseChain: B: PDB Molecule: exosome complex component ski6;PDBTitle: yeast rrp44 nuclease
76 d1mwva1
not modelled
11.6
9
Fold: Heme-dependent peroxidasesSuperfamily: Heme-dependent peroxidasesFamily: Catalase-peroxidase KatG77 c2qz5A_
not modelled
11.2
19
PDB header: signaling protein, lipid binding proteinChain: A: PDB Molecule: axin interactor, dorsalization associatedPDBTitle: crystal structure of the c-terminal domain of aida
78 c3epzA_
not modelled
11.0
26
PDB header: transferaseChain: A: PDB Molecule: dna (cytosine-5)-methyltransferase 1;PDBTitle: structure of the replication foci-targeting sequence of human dna2 cytosine methyltransferase dnmt1
79 d2qalk1
not modelled
11.0
26
Fold: Ribonuclease H-like motifSuperfamily: Translational machinery componentsFamily: Ribosomal protein L18 and S1180 c2br2G_
not modelled
10.9
39
PDB header: hydrolaseChain: G: PDB Molecule: exosome complex exonuclease 2;PDBTitle: rnase ph core of the archaeal exosome
81 d1oysa1
not modelled
10.9
11
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Ribonuclease PH domain 1-like82 c2zkqk_
not modelled
10.9
22
PDB header: ribosomal protein/rnaChain: K: PDB Molecule: PDBTitle: structure of a mammalian ribosomal 40s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
83 c3dd6A_
not modelled
10.6
17
PDB header: transferaseChain: A: PDB Molecule: ribonuclease ph;PDBTitle: crystal structure of rph, an exoribonuclease from bacillus2 anthracis at 1.7 a resolution
84 d2je6a1
not modelled
10.5
39
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Ribonuclease PH domain 1-like85 c2koyA_
not modelled
10.5
12
PDB header: metal transportChain: A: PDB Molecule: copper-transporting atpase 2;PDBTitle: structure of the e1064a mutant of the n-domain of wilson disease2 associated protein
86 d1huwa_
not modelled
10.4
15
Fold: 4-helical cytokinesSuperfamily: 4-helical cytokinesFamily: Long-chain cytokines87 c3bvkC_
not modelled
10.3
13
PDB header: oxidoreductaseChain: C: PDB Molecule: ferritin;PDBTitle: structural basis for the iron uptake mechanism of helicobacter pylori2 ferritin
88 d1ub2a1
not modelled
10.0
12
Fold: Heme-dependent peroxidasesSuperfamily: Heme-dependent peroxidasesFamily: Catalase-peroxidase KatG89 c2arfA_
not modelled
9.9
14
PDB header: hydrolaseChain: A: PDB Molecule: wilson disease atpase;PDBTitle: solution structure of the wilson atpase n-domain in the2 presence of atp
90 d2i09a2
not modelled
9.9
20
Fold: DeoB insert domain-likeSuperfamily: DeoB insert domain-likeFamily: DeoB insert domain-like91 d1v74a_
not modelled
9.5
12
Fold: Colicin D/E5 nuclease domainSuperfamily: Colicin D/E5 nuclease domainFamily: Colicin D nuclease domain92 c3bfjK_
not modelled
9.4
18
PDB header: oxidoreductaseChain: K: PDB Molecule: 1,3-propanediol oxidoreductase;PDBTitle: crystal structure analysis of 1,3-propanediol oxidoreductase
93 d3bzka1
not modelled
9.4
29
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: Tex HhH-containing domain-like94 d1itka1
not modelled
9.4
12
Fold: Heme-dependent peroxidasesSuperfamily: Heme-dependent peroxidasesFamily: Catalase-peroxidase KatG95 c2rq7A_
not modelled
9.3
16
PDB header: hydrolaseChain: A: PDB Molecule: atp synthase epsilon chain;PDBTitle: solution structure of the epsilon subunit chimera combining2 the n-terminal beta-sandwich domain from t. elongatus bp-13 f1 and the c-terminal alpha-helical domain from spinach4 chloroplast f1
96 c2rbbB_
not modelled
9.2
13
PDB header: oxidoreductaseChain: B: PDB Molecule: glyoxalase/bleomycin resistance protein/dioxygenase;PDBTitle: crystal structure of a glyoxalase/bleomycin resistance2 protein/dioxygenase family enzyme from burkholderia phytofirmans psjn
97 c2oudA_
not modelled
9.1
24
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity protein phosphatase 10;PDBTitle: crystal structure of the catalytic domain of human mkp5
98 c2czhB_
not modelled
9.0
33
PDB header: hydrolaseChain: B: PDB Molecule: inositol monophosphatase 2;PDBTitle: crystal structure of human myo-inositol monophosphatase 22 (impa2) with phosphate ion (orthorhombic form)
99 d1ub2a2
not modelled
8.9
15
Fold: Heme-dependent peroxidasesSuperfamily: Heme-dependent peroxidasesFamily: Catalase-peroxidase KatG