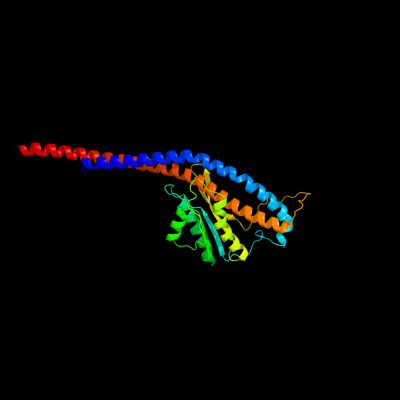
| 1 | c3oaaO_
|
|
 |
100.0 |
100 |
PDB header:hydrolase/transport protein
Chain: O: PDB Molecule:atp synthase gamma chain;
PDBTitle: structure of the e.coli f1-atp synthase inhibited by subunit epsilon
|
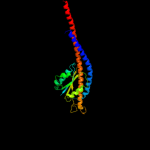
| 2 | c2xokG_
|
|
 |
100.0 |
24 |
PDB header:hydrolase
Chain: G: PDB Molecule:atp synthase subunit gamma, mitochondrial;
PDBTitle: refined structure of yeast f1c10 atpase complex to 3 a2 resolution
|
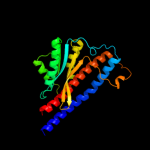
| 3 | d1fs0g_
|
|
 |
100.0 |
99 |
Fold:Pyruvate kinase C-terminal domain-like
Superfamily:ATP synthase (F1-ATPase), gamma subunit
Family:ATP synthase (F1-ATPase), gamma subunit |
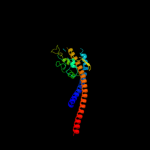
| 4 | d2jdig1
|
|
 |
100.0 |
39 |
Fold:Pyruvate kinase C-terminal domain-like
Superfamily:ATP synthase (F1-ATPase), gamma subunit
Family:ATP synthase (F1-ATPase), gamma subunit |
| 5 | c2qe7G_
|
|
 |
100.0 |
42 |
PDB header:hydrolase
Chain: G: PDB Molecule:atp synthase subunit gamma;
PDBTitle: crystal structure of the f1-atpase from the thermoalkaliphilic2 bacterium bacillus sp. ta2.a1
|
| 6 | c2w6jG_
|
|
 |
100.0 |
41 |
PDB header:hydrolase
Chain: G: PDB Molecule:atp synthase subunit gamma, mitochondrial;
PDBTitle: low resolution structures of bovine mitochondrial f1-atpase2 during controlled dehydration: hydration state 5.
|
| 7 | c3fksY_
|
|
 |
100.0 |
32 |
PDB header:hydrolase
Chain: Y: PDB Molecule:atp synthase subunit gamma, mitochondrial;
PDBTitle: yeast f1 atpase in the absence of bound nucleotides
|
| 8 | d1mabg_
|
|
 |
100.0 |
51 |
Fold:Pyruvate kinase C-terminal domain-like
Superfamily:ATP synthase (F1-ATPase), gamma subunit
Family:ATP synthase (F1-ATPase), gamma subunit |
| 9 | c3re1B_
|
|
 |
75.0 |
8 |
PDB header:lyase
Chain: B: PDB Molecule:uroporphyrinogen-iii synthetase;
PDBTitle: crystal structure of uroporphyrinogen iii synthase from pseudomonas2 syringae pv. tomato dc3000
|
| 10 | d1wd7a_
|
|
 |
61.9 |
18 |
Fold:HemD-like
Superfamily:HemD-like
Family:HemD-like |
| 11 | c3mw8A_
|
|
 |
61.7 |
9 |
PDB header:lyase
Chain: A: PDB Molecule:uroporphyrinogen-iii synthase;
PDBTitle: crystal structure of an uroporphyrinogen-iii synthase (sama_3255) from2 shewanella amazonensis sb2b at 1.65 a resolution
|
| 12 | c3d8tB_
|
|
 |
60.4 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:uroporphyrinogen-iii synthase;
PDBTitle: thermus thermophilus uroporphyrinogen iii synthase
|
| 13 | c2qzuA_
|
|
 |
59.4 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative sulfatase yidj;
PDBTitle: crystal structure of the putative sulfatase yidj from bacteroides2 fragilis. northeast structural genomics consortium target bfr123
|
| 14 | d1fsua_
|
|
 |
54.0 |
15 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:Arylsulfatase |
| 15 | c3b5qB_
|
|
 |
54.0 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative sulfatase yidj;
PDBTitle: crystal structure of a putative sulfatase (np_810509.1)2 from bacteroides thetaiotaomicron vpi-5482 at 2.40 a3 resolution
|
| 16 | d1hdha_
|
|
 |
49.2 |
27 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:Arylsulfatase |
| 17 | c2xr9A_
|
|
 |
46.8 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:ectonucleotide pyrophosphatase/phosphodiesterase family
PDBTitle: crystal structure of autotaxin (enpp2)
|
| 18 | c2kilA_
|
|
 |
44.1 |
15 |
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: nmr structure of the h103g mutant so2144 h-nox domain from2 shewanella oneidensis in the fe(ii)co ligation state
|
| 19 | d1qw2a_
|
|
 |
43.7 |
29 |
Fold:Hypothetical protein Ta1206
Superfamily:Hypothetical protein Ta1206
Family:Hypothetical protein Ta1206 |
| 20 | c3k8vB_
|
|
 |
42.9 |
13 |
PDB header:structural protein
Chain: B: PDB Molecule:flagellin homolog;
PDBTitle: crysatl structure of a bacterial cell-surface flagellin n20c20
|
| 21 | c2xrgA_ |
|
not modelled |
41.8 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:ectonucleotide pyrophosphatase/phosphodiesterase family
PDBTitle: crystal structure of autotaxin (enpp2) in complex with the2 ha155 boronic acid inhibitor
|
| 22 | d1auka_ |
|
not modelled |
39.0 |
11 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:Arylsulfatase |
| 23 | d1p49a_ |
|
not modelled |
37.2 |
17 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:Arylsulfatase |
| 24 | c3p9zA_ |
|
not modelled |
36.5 |
15 |
PDB header:ligase
Chain: A: PDB Molecule:uroporphyrinogen iii cosynthase (hemd);
PDBTitle: crystal structure of uroporphyrinogen-iii synthetase from helicobacter2 pylori 26695
|
| 25 | c2zktB_ |
|
not modelled |
33.9 |
14 |
PDB header:isomerase
Chain: B: PDB Molecule:2,3-bisphosphoglycerate-independent phosphoglycerate
PDBTitle: structure of ph0037 protein from pyrococcus horikoshii
|
| 26 | c3ed4A_ |
|
not modelled |
32.3 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:arylsulfatase;
PDBTitle: crystal structure of putative arylsulfatase from escherichia coli
|
| 27 | c2p1lF_ |
|
not modelled |
32.0 |
39 |
PDB header:apoptosis
Chain: F: PDB Molecule:beclin 1;
PDBTitle: structure of the bcl-xl:beclin 1 complex
|
| 28 | c2w8dB_ |
|
not modelled |
27.6 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:processed glycerol phosphate lipoteichoic acid synthase 2;
PDBTitle: distinct and essential morphogenic functions for wall- and2 lipo-teichoic acids in bacillus subtilis
|
| 29 | c3q3qA_ |
|
not modelled |
27.5 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:alkaline phosphatase;
PDBTitle: crystal structure of spap: an novel alkaline phosphatase from2 bacterium sphingomonas sp. strain bsar-1
|
| 30 | d2pk8a1 |
|
not modelled |
26.8 |
14 |
Fold:Hypothetical protein PF0899
Superfamily:Hypothetical protein PF0899
Family:Hypothetical protein PF0899 |
| 31 | c2vqrA_ |
|
not modelled |
25.9 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative sulfatase;
PDBTitle: crystal structure of a phosphonate monoester hydrolase2 from rhizobium leguminosarum: a new member of the3 alkaline phosphatase superfamily
|
| 32 | c3dvuC_ |
|
not modelled |
24.7 |
39 |
PDB header:viral protein/apoptosis
Chain: C: PDB Molecule:beclin-1;
PDBTitle: crystal structure of the complex of murine gamma-2 herpesvirus 68 bcl-2 homolog m11 and the beclin 1 bh33 domain
|
| 33 | d1sfea2 |
|
not modelled |
24.2 |
42 |
Fold:Ribonuclease H-like motif
Superfamily:Methylated DNA-protein cysteine methyltransferase domain
Family:Methylated DNA-protein cysteine methyltransferase domain |
| 34 | c1o98A_ |
|
not modelled |
24.2 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:2,3-bisphosphoglycerate-independent
PDBTitle: 1.4a crystal structure of phosphoglycerate mutase from2 bacillus stearothermophilus complexed with3 2-phosphoglycerate
|
| 35 | c2i09A_ |
|
not modelled |
23.8 |
12 |
PDB header:isomerase
Chain: A: PDB Molecule:phosphopentomutase;
PDBTitle: crystal structure of putative phosphopentomutase from streptococcus2 mutans
|
| 36 | c3lzcA_ |
|
not modelled |
23.8 |
23 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:dph2;
PDBTitle: crystal structure of dph2 from pyrococcus horikoshii
|
| 37 | d2fefa1 |
|
not modelled |
22.6 |
32 |
Fold:Bromodomain-like
Superfamily:PA2201 N-terminal domain-like
Family:PA2201 N-terminal domain-like |
| 38 | c2w5tA_ |
|
not modelled |
22.6 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:processed glycerol phosphate lipoteichoic acid
PDBTitle: structure-based mechanism of lipoteichoic acid synthesis by2 staphylococcus aureus ltas.
|
| 39 | c3lxqB_ |
|
not modelled |
22.4 |
11 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein vp1736;
PDBTitle: the crystal structure of a protein in the alkaline2 phosphatase superfamily from vibrio parahaemolyticus to3 1.95a
|
| 40 | c2iucB_ |
|
not modelled |
19.7 |
13 |
PDB header:hydrolase
Chain: B: PDB Molecule:alkaline phosphatase;
PDBTitle: structure of alkaline phosphatase from the antarctic2 bacterium tab5
|
| 41 | d1o98a2 |
|
not modelled |
18.2 |
15 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, catalytic domain |
| 42 | c3m8yC_ |
|
not modelled |
17.6 |
10 |
PDB header:isomerase
Chain: C: PDB Molecule:phosphopentomutase;
PDBTitle: phosphopentomutase from bacillus cereus after glucose-1,6-bisphosphate2 activation
|
| 43 | c2ponA_ |
|
not modelled |
17.3 |
39 |
PDB header:apoptosis inhibitor
Chain: A: PDB Molecule:beclin-1;
PDBTitle: solution structure of the bcl-xl/beclin-1 complex
|
| 44 | d1tlla2 |
|
not modelled |
17.1 |
22 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Cytochrome p450 reductase N-terminal domain-like |
| 45 | c3o3xA_ |
|
not modelled |
16.4 |
34 |
PDB header:viral protein
Chain: A: PDB Molecule:gp41-5;
PDBTitle: crystal structure of gp41-5, a single-chain 5-helix-bundle based on2 hiv gp41
|
| 46 | d1kxpd3 |
|
not modelled |
16.2 |
21 |
Fold:Serum albumin-like
Superfamily:Serum albumin-like
Family:Serum albumin-like |
| 47 | d1ewsa_ |
|
not modelled |
14.7 |
50 |
Fold:Defensin-like
Superfamily:Defensin-like
Family:Defensin |
| 48 | c3cseA_ |
|
not modelled |
14.4 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydrofolate reductase;
PDBTitle: candida glabrata dihydrofolate reductase complexed with2 nadph and 2,4-diamino-5-(3-(2,5-dimethoxyphenyl)prop-1-3 ynyl)-6-ethylpyrimidine (ucp120b)
|
| 49 | c3cu2A_ |
|
not modelled |
14.4 |
9 |
PDB header:isomerase
Chain: A: PDB Molecule:ribulose-5-phosphate 3-epimerase;
PDBTitle: crystal structure of ribulose-5-phosphate 3-epimerase (yp_718263.1)2 from haemophilus somnus 129pt at 1.91 a resolution
|
| 50 | d2i09a1 |
|
not modelled |
13.9 |
13 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:DeoB catalytic domain-like |
| 51 | d1u55a_ |
|
not modelled |
13.7 |
14 |
Fold:Ligand-binding domain in the NO signalling and Golgi transport
Superfamily:Ligand-binding domain in the NO signalling and Golgi transport
Family:H-NOX domain |
| 52 | d1q77a_ |
|
not modelled |
13.3 |
19 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:Universal stress protein-like |
| 53 | c2dg2D_ |
|
not modelled |
13.0 |
10 |
PDB header:protein binding
Chain: D: PDB Molecule:apolipoprotein a-i binding protein;
PDBTitle: crystal structure of mouse apolipoprotein a-i binding2 protein
|
| 54 | c2k2dA_ |
|
not modelled |
12.9 |
25 |
PDB header:metal binding protein
Chain: A: PDB Molecule:ring finger and chy zinc finger domain-
PDBTitle: solution nmr structure of c-terminal domain of human pirh2.2 northeast structural genomics consortium (nesg) target ht2c
|
| 55 | c2o0cB_ |
|
not modelled |
12.4 |
15 |
PDB header:signaling protein
Chain: B: PDB Molecule:alr2278 protein;
PDBTitle: crystal structure of the h-nox domain from nostoc sp. pcc 71202 complexed to no
|
| 56 | d2gqfa1 |
|
not modelled |
10.5 |
57 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:HI0933 N-terminal domain-like |
| 57 | c3lmaC_ |
|
not modelled |
10.1 |
15 |
PDB header:membrane protein
Chain: C: PDB Molecule:stage v sporulation protein ad (spovad);
PDBTitle: crystal structure of the stage v sporulation protein ad2 (spovad) from bacillus licheniformis. northeast structural3 genomics consortium target bir6.
|
| 58 | c1jr2A_ |
|
not modelled |
10.0 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:uroporphyrinogen-iii synthase;
PDBTitle: structure of uroporphyrinogen iii synthase
|
| 59 | d1jr2a_ |
|
not modelled |
10.0 |
18 |
Fold:HemD-like
Superfamily:HemD-like
Family:HemD-like |
| 60 | c3k8wA_ |
|
not modelled |
9.9 |
13 |
PDB header:structural protein
Chain: A: PDB Molecule:flagellin homolog;
PDBTitle: crysatl structure of a bacterial cell-surface flagellin n20c45
|
| 61 | d2z06a1 |
|
not modelled |
9.9 |
29 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:TTHA0625-like |
| 62 | d1h3ob_ |
|
not modelled |
9.7 |
15 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:TBP-associated factors, TAFs |
| 63 | d1v10a3 |
|
not modelled |
9.3 |
40 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
| 64 | c1hucC_ |
|
not modelled |
9.3 |
57 |
PDB header:thiol protease
Chain: C: PDB Molecule:cathepsin b;
PDBTitle: the refined 2.15 angstroms x-ray crystal structure of human2 liver cathepsin b: the structural basis for its specificity
|
| 65 | d1yf2a2 |
|
not modelled |
8.8 |
13 |
Fold:DNA methylase specificity domain
Superfamily:DNA methylase specificity domain
Family:Type I restriction modification DNA specificity domain |
| 66 | d1m2da_ |
|
not modelled |
8.8 |
19 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Thioredoxin-like 2Fe-2S ferredoxin |
| 67 | d1jeya2 |
|
not modelled |
8.7 |
10 |
Fold:vWA-like
Superfamily:vWA-like
Family:Ku70 subunit N-terminal domain |
| 68 | c3opyG_ |
|
not modelled |
8.6 |
15 |
PDB header:transferase
Chain: G: PDB Molecule:6-phosphofructo-1-kinase alpha-subunit;
PDBTitle: crystal structure of pichia pastoris phosphofructokinase in the t-2 state
|
| 69 | c1skhA_ |
|
not modelled |
8.4 |
67 |
PDB header:unknown function
Chain: A: PDB Molecule:major prion protein 2;
PDBTitle: n-terminal (1-30) of bovine prion protein
|
| 70 | c2yruA_ |
|
not modelled |
8.4 |
8 |
PDB header:apoptosis
Chain: A: PDB Molecule:steroid receptor rna activator 1;
PDBTitle: solution structure of mouse steroid receptor rna activator2 1 (sra1) protein
|
| 71 | c3mxzA_ |
|
not modelled |
8.2 |
21 |
PDB header:chaperone
Chain: A: PDB Molecule:tubulin-specific chaperone a;
PDBTitle: crystal structure of tubulin folding cofactor a from arabidopsis2 thaliana
|
| 72 | c3opyH_ |
|
not modelled |
8.2 |
18 |
PDB header:transferase
Chain: H: PDB Molecule:6-phosphofructo-1-kinase beta-subunit;
PDBTitle: crystal structure of pichia pastoris phosphofructokinase in the t-2 state
|
| 73 | c3opyB_ |
|
not modelled |
8.2 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:6-phosphofructo-1-kinase beta-subunit;
PDBTitle: crystal structure of pichia pastoris phosphofructokinase in the t-2 state
|
| 74 | c2bpoA_ |
|
not modelled |
8.1 |
12 |
PDB header:reductase
Chain: A: PDB Molecule:nadph-cytochrom p450 reductase;
PDBTitle: crystal structure of the yeast cpr triple mutant: d74g,2 y75f, k78a.
|
| 75 | c3igzB_ |
|
not modelled |
8.0 |
18 |
PDB header:isomerase
Chain: B: PDB Molecule:cofactor-independent phosphoglycerate mutase;
PDBTitle: crystal structures of leishmania mexicana phosphoglycerate2 mutase at low cobalt concentration
|
| 76 | c2ohcB_ |
|
not modelled |
7.9 |
38 |
PDB header:hydrolase
Chain: B: PDB Molecule:trna-splicing endonuclease;
PDBTitle: structural and mutational analysis of trna-intron splicing2 endonuclease from thermoplasma acidophilum dsm1728
|
| 77 | c2cseW_ |
|
not modelled |
7.7 |
45 |
PDB header:virus
Chain: W: PDB Molecule:major core protein lambda 1;
PDBTitle: features of reovirus outer-capsid protein mu1 revealed by2 electron and image reconstruction of the virion at 7.0-a3 resolution
|
| 78 | c3k1qB_ |
|
not modelled |
7.5 |
36 |
PDB header:virus
Chain: B: PDB Molecule:vp3a, the building block protein of inner shell;
PDBTitle: backbone model of an aquareovirus virion by cryo-electron2 microscopy and bioinformatics
|
| 79 | c1ew2A_ |
|
not modelled |
7.4 |
4 |
PDB header:hydrolase
Chain: A: PDB Molecule:phosphatase;
PDBTitle: crystal structure of a human phosphatase
|
| 80 | d1zeda1 |
|
not modelled |
7.4 |
4 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:Alkaline phosphatase |
| 81 | c3d33B_ |
|
not modelled |
7.3 |
27 |
PDB header:unknown function
Chain: B: PDB Molecule:domain of unknown function with an immunoglobulin-like
PDBTitle: crystal structure of a duf3244 family protein with an immunoglobulin-2 like beta-sandwich fold (bvu_0276) from bacteroides vulgatus atcc3 8482 at 1.70 a resolution
|
| 82 | c2oipE_ |
|
not modelled |
7.0 |
14 |
PDB header:transferase, oxidoreductase
Chain: E: PDB Molecule:chain a, crystal structure of dhfr;
PDBTitle: crystal structure of the s290g active site mutant of ts-2 dhfr from cryptosporidium hominis
|
| 83 | c1ihqA_ |
|
not modelled |
7.0 |
33 |
PDB header:de novo protein
Chain: A: PDB Molecule:chimeric peptide glytm1bzip: tropomyosin alpha
PDBTitle: glytm1bzip: a chimeric peptide model of the n-terminus of a2 rat short alpha tropomyosin with the n-terminus encoded by3 exon 1b
|
| 84 | c2i6sA_ |
|
not modelled |
7.0 |
8 |
PDB header:hydrolase
Chain: A: PDB Molecule:complement c2a fragment;
PDBTitle: complement component c2a
|
| 85 | c3k1qC_ |
|
not modelled |
6.9 |
36 |
PDB header:virus
Chain: C: PDB Molecule:vp3b, the building block protein of inner shell;
PDBTitle: backbone model of an aquareovirus virion by cryo-electron2 microscopy and bioinformatics
|
| 86 | c3n39D_ |
|
not modelled |
6.7 |
9 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:protein nrdi;
PDBTitle: ribonucleotide reductase dimanganese(ii)-nrdf from escherichia coli in2 complex with nrdi
|
| 87 | d1y6va1 |
|
not modelled |
6.5 |
12 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:Alkaline phosphatase |
| 88 | d1t70a_ |
|
not modelled |
6.5 |
29 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:DR1281-like |
| 89 | d1fpza_ |
|
not modelled |
6.5 |
14 |
Fold:(Phosphotyrosine protein) phosphatases II
Superfamily:(Phosphotyrosine protein) phosphatases II
Family:Dual specificity phosphatase-like |
| 90 | d1ucua_ |
|
not modelled |
6.5 |
21 |
Fold:Phase 1 flagellin
Superfamily:Phase 1 flagellin
Family:Phase 1 flagellin |
| 91 | c1jpeA_ |
|
not modelled |
6.0 |
43 |
PDB header:electron transport
Chain: A: PDB Molecule:dsbd-alpha;
PDBTitle: crystal structure of dsbd-alpha; the n-terminal domain of2 dsbd
|
| 92 | d1l6pa_ |
|
not modelled |
6.0 |
43 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Thiol:disulfide interchange protein DsbD, N-terminal domain (DsbD-alpha)
Family:Thiol:disulfide interchange protein DsbD, N-terminal domain (DsbD-alpha) |
| 93 | c3rg9A_ |
|
not modelled |
5.9 |
16 |
PDB header:oxidoreductase/oxidoreductase inhibitor
Chain: A: PDB Molecule:bifunctional dihydrofolate reductase-thymidylate synthase;
PDBTitle: trypanosoma brucei dihydrofolate reductase (tbdhfr) in complex with2 wr99210
|
| 94 | d1c0aa2 |
|
not modelled |
5.8 |
13 |
Fold:DCoH-like
Superfamily:GAD domain-like
Family:GAD domain |
| 95 | c2fo1D_ |
|
not modelled |
5.8 |
18 |
PDB header:gene regulation/signalling protein/dna
Chain: D: PDB Molecule:protein lag-3;
PDBTitle: crystal structure of the csl-notch-mastermind ternary2 complex bound to dna
|
| 96 | d2fo1d1 |
|
not modelled |
5.8 |
18 |
Fold:Non-globular all-alpha subunits of globular proteins
Superfamily:Lag-3 N-terminal region
Family:Lag-3 N-terminal region |
| 97 | d1xo1a2 |
|
not modelled |
5.8 |
22 |
Fold:PIN domain-like
Superfamily:PIN domain-like
Family:5' to 3' exonuclease catalytic domain |
| 98 | c2j63B_ |
|
not modelled |
5.7 |
43 |
PDB header:lyase
Chain: B: PDB Molecule:ap-endonuclease;
PDBTitle: crystal structure of ap endonuclease lmap from leishmania2 major
|
| 99 | c2kxoA_ |
|
not modelled |
5.7 |
12 |
PDB header:cell cycle
Chain: A: PDB Molecule:cell division topological specificity factor;
PDBTitle: solution nmr structure of the cell division regulator mine protein2 from neisseria gonorrhoeae
|