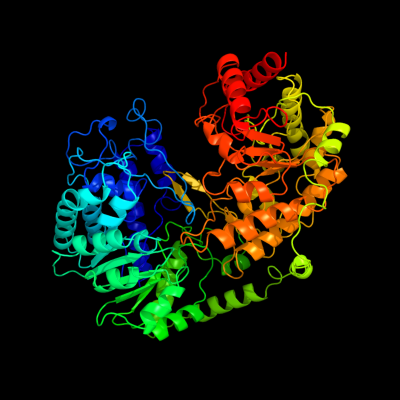
| 1 | c1t7lA_
|
|
 |
100.0 |
42 |
PDB header:transferase
Chain: A: PDB Molecule:5-methyltetrahydropteroyltriglutamate--
PDBTitle: crystal structure of cobalamin-independent methionine2 synthase from t. maritima
|
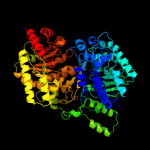
| 2 | c3l7sA_
|
|
 |
100.0 |
46 |
PDB header:transferase
Chain: A: PDB Molecule:5-methyltetrahydropteroyltriglutamate--homocysteine
PDBTitle: crystal structure of mete coordinated with zinc from streptococcus2 mutans
|
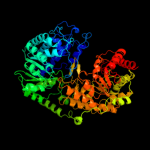
| 3 | c1u22A_
|
|
 |
100.0 |
50 |
PDB header:transferase
Chain: A: PDB Molecule:5-methyltetrahydropteroyltriglutamate--
PDBTitle: a. thaliana cobalamine independent methionine synthase
|
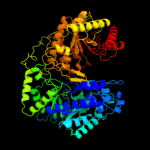
| 4 | c2nq5A_
|
|
 |
100.0 |
46 |
PDB header:transferase
Chain: A: PDB Molecule:5-methyltetrahydropteroyltriglutamate--
PDBTitle: crystal structure of methyltransferase from streptococcus2 mutans
|
| 5 | d1u1ha1
|
|
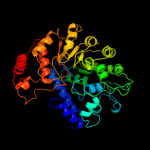 |
100.0 |
41 |
Fold:TIM beta/alpha-barrel
Superfamily:UROD/MetE-like
Family:Cobalamin-independent methionine synthase |
| 6 | d1u1ha2
|
|
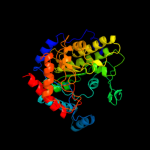 |
100.0 |
58 |
Fold:TIM beta/alpha-barrel
Superfamily:UROD/MetE-like
Family:Cobalamin-independent methionine synthase |
| 7 | c3rpdB_
|
|
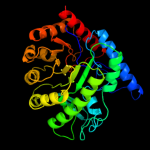 |
100.0 |
27 |
PDB header:transferase
Chain: B: PDB Molecule:methionine synthase (b12-independent);
PDBTitle: the structure of a b12-independent methionine synthase from shewanella2 sp. w3-18-1 in complex with selenomethionine.
|
| 8 | c1ypxA_
|
|
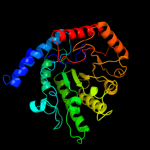 |
100.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:putative vitamin-b12 independent methionine synthase family
PDBTitle: crystal structure of the putative vitamin-b12 independent methionine2 synthase from listeria monocytogenes, northeast structural genomics3 target lmr13
|
| 9 | d1j93a_
|
|
 |
99.6 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:UROD/MetE-like
Family:Uroporphyrinogen decarboxylase, UROD |
| 10 | c3cyvA_
|
|
 |
99.5 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: crystal structure of uroporphyrinogen decarboxylase from2 shigella flexineri: new insights into its catalytic3 mechanism
|
| 11 | c1jpkA_
|
|
 |
99.5 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: gly156asp mutant of human urod, human uroporphyrinogen iii2 decarboxylase
|
| 12 | d1r3sa_
|
|
 |
99.4 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:UROD/MetE-like
Family:Uroporphyrinogen decarboxylase, UROD |
| 13 | c2infB_
|
|
 |
99.4 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: crystal structure of uroporphyrinogen decarboxylase from2 bacillus subtilis
|
| 14 | c2ejaB_
|
|
 |
99.4 |
18 |
PDB header:lyase
Chain: B: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: crystal structure of uroporphyrinogen decarboxylase from2 aquifex aeolicus
|
| 15 | c3qc3B_
|
|
 |
93.7 |
15 |
PDB header:isomerase
Chain: B: PDB Molecule:d-ribulose-5-phosphate-3-epimerase;
PDBTitle: crystal structure of a d-ribulose-5-phosphate-3-epimerase (np_954699)2 from homo sapiens at 2.20 a resolution
|
| 16 | d1rpxa_
|
|
 |
93.5 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
| 17 | d1h1ya_
|
|
 |
91.7 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
| 18 | c3inpA_
|
|
 |
91.7 |
11 |
PDB header:isomerase
Chain: A: PDB Molecule:d-ribulose-phosphate 3-epimerase;
PDBTitle: 2.05 angstrom resolution crystal structure of d-ribulose-phosphate 3-2 epimerase from francisella tularensis.
|
| 19 | d1tqja_
|
|
 |
91.1 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
| 20 | c1yadD_
|
|
 |
90.4 |
8 |
PDB header:transcription
Chain: D: PDB Molecule:regulatory protein teni;
PDBTitle: structure of teni from bacillus subtilis
|
| 21 | c3nm3D_ |
|
not modelled |
90.3 |
18 |
PDB header:transferase
Chain: D: PDB Molecule:thiamine biosynthetic bifunctional enzyme;
PDBTitle: the crystal structure of candida glabrata thi6, a bifunctional enzyme2 involved in thiamin biosyhthesis of eukaryotes
|
| 22 | c3ceuA_ |
|
not modelled |
90.3 |
8 |
PDB header:transferase
Chain: A: PDB Molecule:thiamine phosphate pyrophosphorylase;
PDBTitle: crystal structure of thiamine phosphate pyrophosphorylase2 (bt_0647) from bacteroides thetaiotaomicron. northeast3 structural genomics consortium target btr268
|
| 23 | d1tqxa_ |
|
not modelled |
88.0 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
| 24 | d2tpsa_ |
|
not modelled |
87.7 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Thiamin phosphate synthase
Family:Thiamin phosphate synthase |
| 25 | c3bolB_ |
|
not modelled |
84.6 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:5-methyltetrahydrofolate s-homocysteine
PDBTitle: cobalamin-dependent methionine synthase (1-566) from2 thermotoga maritima complexed with zn2+
|
| 26 | c3cu2A_ |
|
not modelled |
80.7 |
19 |
PDB header:isomerase
Chain: A: PDB Molecule:ribulose-5-phosphate 3-epimerase;
PDBTitle: crystal structure of ribulose-5-phosphate 3-epimerase (yp_718263.1)2 from haemophilus somnus 129pt at 1.91 a resolution
|
| 27 | c3lrmB_ |
|
not modelled |
80.2 |
13 |
PDB header:hydrolase
Chain: B: PDB Molecule:alpha-galactosidase 1;
PDBTitle: structure of alfa-galactosidase from saccharomyces cerevisiae with2 raffinose
|
| 28 | d1xi3a_ |
|
not modelled |
79.4 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Thiamin phosphate synthase
Family:Thiamin phosphate synthase |
| 29 | c3ipwA_ |
|
not modelled |
76.3 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:hydrolase tatd family protein;
PDBTitle: crystal structure of hydrolase tatd family protein from entamoeba2 histolytica
|
| 30 | d1ojxa_ |
|
not modelled |
75.8 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 31 | c3a23A_ |
|
not modelled |
75.5 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative secreted alpha-galactosidase;
PDBTitle: crystal structure of beta-l-arabinopyranosidase complexed with d-2 galactose
|
| 32 | c2ze3A_ |
|
not modelled |
71.9 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:dfa0005;
PDBTitle: crystal structure of dfa0005 complexed with alpha-ketoglutarate: a2 novel member of the icl/pepm superfamily from alkali-tolerant3 deinococcus ficus
|
| 33 | c2bmbA_ |
|
not modelled |
70.9 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:folic acid synthesis protein fol1;
PDBTitle: x-ray structure of the bifunctional 6-hydroxymethyl-7,8-2 dihydroxypterin pyrophosphokinase dihydropteroate synthase3 from saccharomyces cerevisiae
|
| 34 | d3bofa2 |
|
not modelled |
69.2 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Homocysteine S-methyltransferase
Family:Homocysteine S-methyltransferase |
| 35 | c2h90A_ |
|
not modelled |
69.1 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:xenobiotic reductase a;
PDBTitle: xenobiotic reductase a in complex with coumarin
|
| 36 | d1ur4a_ |
|
not modelled |
63.3 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 37 | d1r46a2 |
|
not modelled |
60.8 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 38 | d2flia1 |
|
not modelled |
60.4 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
| 39 | d1uasa2 |
|
not modelled |
58.8 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 40 | d1qapa1 |
|
not modelled |
57.9 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Nicotinate/Quinolinate PRTase C-terminal domain-like
Family:NadC C-terminal domain-like |
| 41 | c1tx2A_ |
|
not modelled |
57.0 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:dhps, dihydropteroate synthase;
PDBTitle: dihydropteroate synthetase, with bound inhibitor manic, from bacillus2 anthracis
|
| 42 | d1tx2a_ |
|
not modelled |
57.0 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 43 | c3ct7E_ |
|
not modelled |
56.6 |
10 |
PDB header:isomerase
Chain: E: PDB Molecule:d-allulose-6-phosphate 3-epimerase;
PDBTitle: crystal structure of d-allulose 6-phosphate 3-epimerase2 from escherichia coli k-12
|
| 44 | c2qjhH_ |
|
not modelled |
56.1 |
17 |
PDB header:lyase
Chain: H: PDB Molecule:putative aldolase mj0400;
PDBTitle: m. jannaschii adh synthase covalently bound to2 dihydroxyacetone phosphate
|
| 45 | c3tr9A_ |
|
not modelled |
56.1 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: structure of a dihydropteroate synthase (folp) in complex with pteroic2 acid from coxiella burnetii
|
| 46 | d1ps9a1 |
|
not modelled |
55.3 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 47 | c2jbmA_ |
|
not modelled |
54.3 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:nicotinate-nucleotide pyrophosphorylase;
PDBTitle: qprtase structure from human
|
| 48 | c1j0yD_ |
|
not modelled |
53.6 |
14 |
PDB header:hydrolase
Chain: D: PDB Molecule:beta-amylase;
PDBTitle: beta-amylase from bacillus cereus var. mycoides in complex2 with glucose
|
| 49 | c1qapA_ |
|
not modelled |
53.6 |
20 |
PDB header:glycosyltransferase
Chain: A: PDB Molecule:quinolinic acid phosphoribosyltransferase;
PDBTitle: quinolinic acid phosphoribosyltransferase with bound2 quinolinic acid
|
| 50 | c3l2iB_ |
|
not modelled |
53.5 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: 1.85 angstrom crystal structure of the 3-dehydroquinate dehydratase2 (arod) from salmonella typhimurium lt2.
|
| 51 | c1ps9A_ |
|
not modelled |
52.7 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2,4-dienoyl-coa reductase;
PDBTitle: the crystal structure and reaction mechanism of e. coli 2,4-2 dienoyl coa reductase
|
| 52 | c3a5vA_ |
|
not modelled |
52.3 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-galactosidase;
PDBTitle: crystal structure of alpha-galactosidase i from mortierella vinacea
|
| 53 | d1vema2 |
|
not modelled |
51.9 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 54 | c2gl0A_ |
|
not modelled |
51.5 |
32 |
PDB header:transferase
Chain: A: PDB Molecule:conserved hypothetical protein;
PDBTitle: structure of pae2307 in complex with adenosine
|
| 55 | c3hf3A_ |
|
not modelled |
50.4 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:chromate reductase;
PDBTitle: old yellow enzyme from thermus scotoductus sa-01
|
| 56 | c1o4uA_ |
|
not modelled |
47.1 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:type ii quinolic acid phosphoribosyltransferase;
PDBTitle: crystal structure of a nicotinate nucleotide pyrophosphorylase2 (tm1645) from thermotoga maritima at 2.50 a resolution
|
| 57 | c3e2vA_ |
|
not modelled |
45.7 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:3'-5'-exonuclease;
PDBTitle: crystal structure of an uncharacterized amidohydrolase from2 saccharomyces cerevisiae
|
| 58 | c2y5sA_ |
|
not modelled |
44.6 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: crystal structure of burkholderia cenocepacia dihydropteroate2 synthase complexed with 7,8-dihydropteroate.
|
| 59 | c2d16B_ |
|
not modelled |
43.4 |
21 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical protein ph1918;
PDBTitle: crystal structure of ph1918 protein from pyrococcus horikoshii ot3
|
| 60 | c1x1oC_ |
|
not modelled |
42.7 |
21 |
PDB header:transferase
Chain: C: PDB Molecule:nicotinate-nucleotide pyrophosphorylase;
PDBTitle: crystal structure of project id tt0268 from thermus thermophilus hb8
|
| 61 | d1e0ta2 |
|
not modelled |
42.4 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 62 | d1o4ua1 |
|
not modelled |
41.7 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Nicotinate/Quinolinate PRTase C-terminal domain-like
Family:NadC C-terminal domain-like |
| 63 | d1vgga_ |
|
not modelled |
41.0 |
22 |
Fold:Ta1353-like
Superfamily:Ta1353-like
Family:Ta1353-like |
| 64 | c3f41B_ |
|
not modelled |
40.7 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:phytase;
PDBTitle: structure of the tandemly repeated protein tyrosine2 phosphatase like phytase from mitsuokella multacida
|
| 65 | d1rd5a_ |
|
not modelled |
40.3 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 66 | c1djnB_ |
|
not modelled |
40.1 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:trimethylamine dehydrogenase;
PDBTitle: structural and biochemical characterization of recombinant wild type2 trimethylamine dehydrogenase from methylophilus methylotrophus (sp.3 w3a1)
|
| 67 | c2jvfA_ |
|
not modelled |
39.8 |
20 |
PDB header:de novo protein
Chain: A: PDB Molecule:de novo protein m7;
PDBTitle: solution structure of m7, a computationally-designed2 artificial protein
|
| 68 | d1qopa_ |
|
not modelled |
39.6 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 69 | c3bg3B_ |
|
not modelled |
37.9 |
12 |
PDB header:ligase
Chain: B: PDB Molecule:pyruvate carboxylase, mitochondrial;
PDBTitle: crystal structure of human pyruvate carboxylase (missing2 the biotin carboxylase domain at the n-terminus)
|
| 70 | d1xcfa_ |
|
not modelled |
37.7 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 71 | c3thaB_ |
|
not modelled |
35.1 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: tryptophan synthase subunit alpha from campylobacter jejuni.
|
| 72 | c3l0gD_ |
|
not modelled |
35.1 |
16 |
PDB header:transferase
Chain: D: PDB Molecule:nicotinate-nucleotide pyrophosphorylase;
PDBTitle: crystal structure of nicotinate-nucleotide pyrophosphorylase from2 ehrlichia chaffeensis at 2.05a resolution
|
| 73 | d1ad1a_ |
|
not modelled |
34.9 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 74 | c3labA_ |
|
not modelled |
34.6 |
12 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative kdpg (2-keto-3-deoxy-6-phosphogluconate)
PDBTitle: crystal structure of a putative kdpg (2-keto-3-deoxy-6-2 phosphogluconate) aldolase from oleispira antarctica
|
| 75 | d1luca_ |
|
not modelled |
34.3 |
7 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Bacterial luciferase (alkanal monooxygenase) |
| 76 | c2b7pA_ |
|
not modelled |
34.3 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:probable nicotinate-nucleotide pyrophosphorylase;
PDBTitle: crystal structure of quinolinic acid phosphoribosyltransferase from2 helicobacter pylori
|
| 77 | d1umya_ |
|
not modelled |
33.1 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Homocysteine S-methyltransferase
Family:Homocysteine S-methyltransferase |
| 78 | c2xioA_ |
|
not modelled |
32.6 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative deoxyribonuclease tatdn1;
PDBTitle: structure of putative deoxyribonuclease tatdn1 isoform a
|
| 79 | c2fcdA_ |
|
not modelled |
31.0 |
13 |
PDB header:cell cycle
Chain: A: PDB Molecule:myosin light chain 1;
PDBTitle: solution structure of n-lobe myosin light chain from2 saccharomices cerevisiae
|
| 80 | c3pajA_ |
|
not modelled |
30.7 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:nicotinate-nucleotide pyrophosphorylase, carboxylating;
PDBTitle: 2.00 angstrom resolution crystal structure of a quinolinate2 phosphoribosyltransferase from vibrio cholerae o1 biovar eltor str.3 n16961
|
| 81 | c1mh3A_ |
|
not modelled |
29.8 |
12 |
PDB header:sugar binding, dna binding protein
Chain: A: PDB Molecule:maltose binding-a1 homeodomain protein chimera;
PDBTitle: maltose binding-a1 homeodomain protein chimera, crystal2 form i
|
| 82 | c3js3C_ |
|
not modelled |
28.8 |
13 |
PDB header:lyase
Chain: C: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: crystal structure of type i 3-dehydroquinate dehydratase (arod) from2 clostridium difficile with covalent reaction intermediate
|
| 83 | d3bofa1 |
|
not modelled |
28.3 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Methyltetrahydrofolate-utiluzing methyltransferases |
| 84 | c3o63B_ |
|
not modelled |
27.4 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:probable thiamine-phosphate pyrophosphorylase;
PDBTitle: crystal structure of thiamin phosphate synthase from mycobacterium2 tuberculosis
|
| 85 | c3fpjA_ |
|
not modelled |
27.4 |
16 |
PDB header:biosynthetic protein, transferase
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of e81q mutant of mtnas in complex with s-2 adenosylmethionine
|
| 86 | c2wdfA_ |
|
not modelled |
26.9 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:sulfur oxidation protein soxb;
PDBTitle: termus thermophilus sulfate thiohydrolase soxb
|
| 87 | d1gqna_ |
|
not modelled |
26.7 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 88 | d1wa3a1 |
|
not modelled |
26.5 |
7 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 89 | c1qysA_ |
|
not modelled |
24.4 |
13 |
PDB header:de novo protein
Chain: A: PDB Molecule:top7;
PDBTitle: crystal structure of top7: a computationally designed2 protein with a novel fold
|
| 90 | c3l5aA_ |
|
not modelled |
24.2 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadh/flavin oxidoreductase/nadh oxidase;
PDBTitle: crystal structure of a probable nadh-dependent flavin oxidoreductase2 from staphylococcus aureus
|
| 91 | c3rhgA_ |
|
not modelled |
23.8 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative phophotriesterase;
PDBTitle: crystal structure of amidohydrolase pmi1525 (target efi-500319) from2 proteus mirabilis hi4320
|
| 92 | c2v82A_ |
|
not modelled |
23.7 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:2-dehydro-3-deoxy-6-phosphogalactonate aldolase;
PDBTitle: kdpgal complexed to kdpgal
|
| 93 | c2ekcA_ |
|
not modelled |
23.4 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: structural study of project id aq_1548 from aquifex aeolicus vf5
|
| 94 | c3a98A_ |
|
not modelled |
23.3 |
16 |
PDB header:signaling protein
Chain: A: PDB Molecule:dedicator of cytokinesis protein 2;
PDBTitle: crystal structure of the complex of the interacting regions of dock22 and elmo1
|
| 95 | d1pkla2 |
|
not modelled |
23.1 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 96 | c2yr1B_ |
|
not modelled |
23.1 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: crystal structure of 3-dehydroquinate dehydratase from geobacillus2 kaustophilus hta426
|
| 97 | d1vjia_ |
|
not modelled |
22.9 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 98 | d1piia1 |
|
not modelled |
22.4 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 99 | d1ajza_ |
|
not modelled |
21.7 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 100 | c3gndC_ |
|
not modelled |
21.7 |
15 |
PDB header:lyase
Chain: C: PDB Molecule:aldolase lsrf;
PDBTitle: crystal structure of e. coli lsrf in complex with ribulose-5-phosphate
|
| 101 | d1qpoa1 |
|
not modelled |
21.4 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Nicotinate/Quinolinate PRTase C-terminal domain-like
Family:NadC C-terminal domain-like |
| 102 | c2vefB_ |
|
not modelled |
21.3 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:dihydropteroate synthase;
PDBTitle: dihydropteroate synthase from streptococcus pneumoniae
|