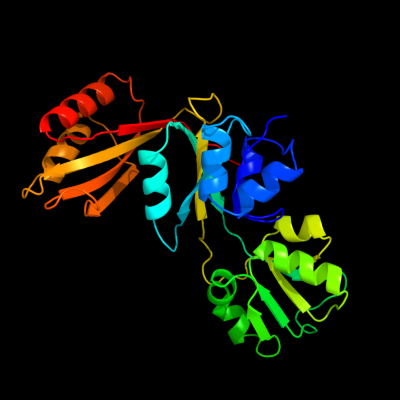
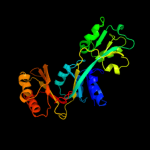
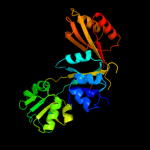
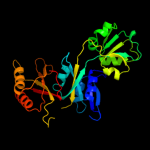
1 c3mwbA_
100.0
25
PDB header: lyaseChain: A: PDB Molecule: prephenate dehydratase;PDBTitle: the crystal structure of prephenate dehydratase in complex with l-phe2 from arthrobacter aurescens to 2.0a
2 c2qmxB_
100.0
33
PDB header: ligaseChain: B: PDB Molecule: prephenate dehydratase;PDBTitle: the crystal structure of l-phe inhibited prephenate dehydratase from2 chlorobium tepidum tls
3 c3luyA_
100.0
23
PDB header: isomeraseChain: A: PDB Molecule: probable chorismate mutase;PDBTitle: putative chorismate mutase from bifidobacterium adolescentis
4 c2qmwA_
100.0
25
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: prephenate dehydratase;PDBTitle: the crystal structure of the prephenate dehydratase (pdt) from2 staphylococcus aureus subsp. aureus mu50
5 d2qmwa1
100.0
28
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like6 d2qmwa2
99.9
17
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Phenylalanine metabolism regulatory domain7 d1phza1
99.9
20
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Phenylalanine metabolism regulatory domain8 c3rmiA_
99.8
26
PDB header: isomeraseChain: A: PDB Molecule: chorismate mutase protein;PDBTitle: crystal structure of chorismate mutase from bartonella henselae str.2 houston-1 in complex with malate
9 d1ecma_
99.8
100
Fold: Chorismate mutase IISuperfamily: Chorismate mutase IIFamily: Dimeric chorismate mutase10 d2gtvx1
99.8
34
Fold: Chorismate mutase IISuperfamily: Chorismate mutase IIFamily: monomeric chorismate mutase11 d2d8da1
99.8
28
Fold: Chorismate mutase IISuperfamily: Chorismate mutase IIFamily: Dimeric chorismate mutase12 c3nvtA_
99.7
25
PDB header: transferase/isomeraseChain: A: PDB Molecule: 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase;PDBTitle: 1.95 angstrom crystal structure of a bifunctional 3-deoxy-7-2 phosphoheptulonate synthase/chorismate mutase (aroa) from listeria3 monocytogenes egd-e
13 d1ybza1
99.7
33
Fold: Chorismate mutase IISuperfamily: Chorismate mutase IIFamily: Dimeric chorismate mutase14 d2fp1a1
99.6
23
Fold: Chorismate mutase IISuperfamily: Chorismate mutase IIFamily: Secreted chorismate mutase-like15 d2h9da1
99.6
22
Fold: Chorismate mutase IISuperfamily: Chorismate mutase IIFamily: Dimeric chorismate mutase16 c2gbbA_
99.6
19
PDB header: isomeraseChain: A: PDB Molecule: putative chorismate mutase;PDBTitle: crystal structure of secreted chorismate mutase from2 yersinia pestis
17 c2qbvA_
99.5
33
PDB header: isomeraseChain: A: PDB Molecule: chorismate mutase;PDBTitle: crystal structure of intracellular chorismate mutase from2 mycobacterium tuberculosis
18 c2phmA_
99.3
21
PDB header: oxidoreductaseChain: A: PDB Molecule: protein (phenylalanine-4-hydroxylase);PDBTitle: structure of phenylalanine hydroxylase dephosphorylated
19 d1ygya3
97.4
13
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Phosphoglycerate dehydrogenase, regulatory (C-terminal) domain20 c1u8sB_
97.4
9
PDB header: transcriptionChain: B: PDB Molecule: glycine cleavage system transcriptionalPDBTitle: crystal structure of putative glycine cleavage system2 transcriptional repressor
21 c2nyiB_
not modelled
97.2
11
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: unknown protein;PDBTitle: crystal structure of an unknown protein from galdieria2 sulphuraria
22 c3ibwA_
not modelled
97.1
6
PDB header: transferaseChain: A: PDB Molecule: gtp pyrophosphokinase;PDBTitle: crystal structure of the act domain from gtp2 pyrophosphokinase of chlorobium tepidum. northeast3 structural genomics consortium target ctr148a
23 d2f1fa1
not modelled
97.0
14
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: IlvH-like24 d1u8sa2
not modelled
96.9
9
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Glycine cleavage system transcriptional repressor25 d2pc6a2
not modelled
96.9
15
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: IlvH-like26 c2f1fA_
not modelled
96.8
14
PDB header: transferaseChain: A: PDB Molecule: acetolactate synthase isozyme iii small subunit;PDBTitle: crystal structure of the regulatory subunit of2 acetohydroxyacid synthase isozyme iii from e. coli
27 d2f06a1
not modelled
96.6
18
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: BT0572-like28 d2fgca2
not modelled
96.5
7
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: IlvH-like29 d5csma_
not modelled
96.5
22
Fold: Chorismate mutase IISuperfamily: Chorismate mutase IIFamily: Allosteric chorismate mutase30 c2pc6C_
not modelled
96.4
15
PDB header: lyaseChain: C: PDB Molecule: probable acetolactate synthase isozyme iii (small subunit);PDBTitle: crystal structure of putative acetolactate synthase- small subunit2 from nitrosomonas europaea
31 d1zpva1
not modelled
96.4
4
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: SP0238-like32 d2f06a2
not modelled
96.0
21
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: BT0572-like33 d1sc6a3
not modelled
96.0
15
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Phosphoglycerate dehydrogenase, regulatory (C-terminal) domain34 c2fgcA_
not modelled
96.0
7
PDB header: transferaseChain: A: PDB Molecule: acetolactate synthase, small subunit;PDBTitle: crystal structure of acetolactate synthase- small subunit from2 thermotoga maritima
35 c1ygyA_
not modelled
95.9
11
PDB header: oxidoreductaseChain: A: PDB Molecule: d-3-phosphoglycerate dehydrogenase;PDBTitle: crystal structure of d-3-phosphoglycerate dehydrogenase from2 mycobacterium tuberculosis
36 c2f06B_
not modelled
95.3
20
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: conserved hypothetical protein;PDBTitle: crystal structure of protein bt0572 from bacteroides thetaiotaomicron
37 d1u8sa1
not modelled
94.9
9
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Glycine cleavage system transcriptional repressor38 c3n0vD_
not modelled
94.7
13
PDB header: hydrolaseChain: D: PDB Molecule: formyltetrahydrofolate deformylase;PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (pp_0327)2 from pseudomonas putida kt2440 at 2.25 a resolution
39 c1y7pB_
not modelled
94.5
13
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical protein af1403;PDBTitle: 1.9 a crystal structure of a protein of unknown function2 af1403 from archaeoglobus fulgidus, probable metabolic3 regulator
40 c3l6gA_
not modelled
94.1
17
PDB header: glycine betaine-binding proteinChain: A: PDB Molecule: betaine abc transporter permease and substrate bindingPDBTitle: crystal structure of lactococcal opuac in its open conformation
41 c1tdjA_
not modelled
94.0
14
PDB header: allosteryChain: A: PDB Molecule: biosynthetic threonine deaminase;PDBTitle: threonine deaminase (biosynthetic) from e. coli
42 c3o1lB_
not modelled
93.8
17
PDB header: hydrolaseChain: B: PDB Molecule: formyltetrahydrofolate deformylase;PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (pspto_4314)2 from pseudomonas syringae pv. tomato str. dc3000 at 2.20 a resolution
43 c3louB_
not modelled
93.1
14
PDB header: hydrolaseChain: B: PDB Molecule: formyltetrahydrofolate deformylase;PDBTitle: crystal structure of formyltetrahydrofolate deformylase (yp_105254.1)2 from burkholderia mallei atcc 23344 at 1.90 a resolution
44 c3mtjA_
not modelled
92.8
11
PDB header: oxidoreductaseChain: A: PDB Molecule: homoserine dehydrogenase;PDBTitle: the crystal structure of a homoserine dehydrogenase from thiobacillus2 denitrificans to 2.15a
45 d1xs5a_
not modelled
92.5
7
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like46 c3gxaA_
not modelled
92.2
11
PDB header: protein bindingChain: A: PDB Molecule: outer membrane lipoprotein gna1946;PDBTitle: crystal structure of gna1946
47 c3tmgA_
not modelled
91.9
13
PDB header: transport proteinChain: A: PDB Molecule: glycine betaine, l-proline abc transporter,PDBTitle: crystal structure of glycine betaine, l-proline abc transporter,2 glycine/betaine/l-proline-binding protein (prox) from borrelia3 burgdorferi
48 c3l76B_
not modelled
91.7
10
PDB header: transferaseChain: B: PDB Molecule: aspartokinase;PDBTitle: crystal structure of aspartate kinase from synechocystis
49 c2f5xC_
not modelled
91.5
16
PDB header: transport proteinChain: C: PDB Molecule: bugd;PDBTitle: structure of periplasmic binding protein bugd
50 c3tqwA_
not modelled
89.8
11
PDB header: transport proteinChain: A: PDB Molecule: methionine-binding protein;PDBTitle: structure of a abc transporter, periplasmic substrate-binding protein2 from coxiella burnetii
51 c3nrbD_
not modelled
89.7
13
PDB header: hydrolaseChain: D: PDB Molecule: formyltetrahydrofolate deformylase;PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (puru,2 pp_1943) from pseudomonas putida kt2440 at 2.05 a resolution
52 c3ir1F_
not modelled
89.0
11
PDB header: protein bindingChain: F: PDB Molecule: outer membrane lipoprotein gna1946;PDBTitle: crystal structure of lipoprotein gna1946 from neisseria2 meningitidis
53 c3k2dA_
not modelled
88.4
5
PDB header: immune systemChain: A: PDB Molecule: abc-type metal ion transport system, periplasmic component;PDBTitle: crystal structure of immunogenic lipoprotein a from vibrio vulnificus
54 c1p99A_
not modelled
88.0
8
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein pg110;PDBTitle: 1.7a crystal structure of protein pg110 from staphylococcus2 aureus
55 d1p99a_
not modelled
88.0
8
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like56 c3e4rA_
not modelled
87.8
14
PDB header: transport proteinChain: A: PDB Molecule: nitrate transport protein;PDBTitle: crystal structure of the alkanesulfonate binding protein2 (ssua) from the phytopathogenic bacteria xanthomonas3 axonopodis pv. citri bound to hepes
57 c2dvzA_
not modelled
86.9
14
PDB header: transport proteinChain: A: PDB Molecule: putative exported protein;PDBTitle: structure of a periplasmic transporter
58 c2x26A_
not modelled
84.8
11
PDB header: transport proteinChain: A: PDB Molecule: periplasmic aliphatic sulphonates-binding protein;PDBTitle: crystal structure of the periplasmic aliphatic sulphonate2 binding protein ssua from escherichia coli
59 c2qpqC_
not modelled
84.5
17
PDB header: transport proteinChain: C: PDB Molecule: protein bug27;PDBTitle: structure of bug27 from bordetella pertussis
60 c3obiC_
not modelled
84.2
11
PDB header: hydrolaseChain: C: PDB Molecule: formyltetrahydrofolate deformylase;PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (np_949368)2 from rhodopseudomonas palustris cga009 at 1.95 a resolution
61 d1tdja2
not modelled
83.7
13
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Allosteric threonine deaminase C-terminal domain62 c1r48A_
not modelled
83.1
26
PDB header: transport proteinChain: A: PDB Molecule: proline/betaine transporter;PDBTitle: solution structure of the c-terminal cytoplasmic domain2 residues 468-497 of escherichia coli protein prop
63 c1u8cB_
not modelled
83.1
16
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-3;PDBTitle: a novel adaptation of the integrin psi domain revealed from its2 crystal structure
64 c2dtjA_
not modelled
81.8
10
PDB header: transferaseChain: A: PDB Molecule: aspartokinase;PDBTitle: crystal structure of regulatory subunit of aspartate kinase2 from corynebacterium glutamicum
65 c3un6A_
not modelled
80.9
10
PDB header: unknown functionChain: A: PDB Molecule: hypothetical protein saouhsc_00137;PDBTitle: 2.0 angstrom crystal structure of ligand binding component of abc-type2 import system from staphylococcus aureus with zinc bound
66 c3n5lA_
not modelled
79.8
15
PDB header: transport proteinChain: A: PDB Molecule: binding protein component of abc phosphonate transporter;PDBTitle: crystal structure of a binding protein component of abc phosphonate2 transporter (pa3383) from pseudomonas aeruginosa at 1.97 a resolution
67 c1ybaC_
not modelled
77.6
15
PDB header: oxidoreductaseChain: C: PDB Molecule: d-3-phosphoglycerate dehydrogenase;PDBTitle: the active form of phosphoglycerate dehydrogenase
68 c3k6sB_
not modelled
76.7
23
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-2;PDBTitle: structure of integrin alphaxbeta2 ectodomain
69 c2de4B_
not modelled
74.1
8
PDB header: hydrolaseChain: B: PDB Molecule: dibenzothiophene desulfurization enzyme b;PDBTitle: crystal structure of dszb c27s mutant in complex with biphenyl-2-2 sulfinic acid
70 c3fcuB_
not modelled
73.8
16
PDB header: cell adhesion/blood clottingChain: B: PDB Molecule: integrin beta-3;PDBTitle: structure of headpiece of integrin aiibb3 in open conformation
71 c3uifA_
not modelled
72.7
19
PDB header: transport proteinChain: A: PDB Molecule: sulfonate abc transporter, periplasmic sulfonate-bindingPDBTitle: crystal structure of putative sulfonate abc transporter, periplasmic2 sulfonate-binding protein ssua from methylobacillus flagellatus kt
72 c3hn0A_
not modelled
72.6
9
PDB header: transport proteinChain: A: PDB Molecule: nitrate transport protein;PDBTitle: crystal structure of an abc transporter (bdi_1369) from2 parabacteroides distasonis at 1.75 a resolution
73 c2zhoB_
not modelled
69.1
13
PDB header: transferaseChain: B: PDB Molecule: aspartokinase;PDBTitle: crystal structure of the regulatory subunit of aspartate2 kinase from thermus thermophilus (ligand free form)
74 c3qslA_
not modelled
69.0
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative exported protein;PDBTitle: structure of cae31940 from bordetella bronchiseptica rb50
75 c2j0wA_
not modelled
66.5
16
PDB header: transferaseChain: A: PDB Molecule: lysine-sensitive aspartokinase 3;PDBTitle: crystal structure of e. coli aspartokinase iii in complex2 with aspartate and adp (r-state)
76 c3ix1A_
not modelled
66.0
10
PDB header: biosynthetic proteinChain: A: PDB Molecule: n-formyl-4-amino-5-aminomethyl-2-methylpyrimidine bindingPDBTitle: periplasmic n-formyl-4-amino-5-aminomethyl-2-methylpyrimidine binding2 protein from bacillus halodurans
77 c3ix1B_
not modelled
66.0
10
PDB header: biosynthetic proteinChain: B: PDB Molecule: n-formyl-4-amino-5-aminomethyl-2-methylpyrimidine bindingPDBTitle: periplasmic n-formyl-4-amino-5-aminomethyl-2-methylpyrimidine binding2 protein from bacillus halodurans
78 c3ab4K_
not modelled
65.5
10
PDB header: transferaseChain: K: PDB Molecule: aspartokinase;PDBTitle: crystal structure of feedback inhibition resistant mutant of aspartate2 kinase from corynebacterium glutamicum in complex with lysine and3 threonine
79 c3p96A_
not modelled
65.5
25
PDB header: hydrolaseChain: A: PDB Molecule: phosphoserine phosphatase serb;PDBTitle: crystal structure of phosphoserine phosphatase serb from mycobacterium2 avium, native form
80 d1tdja3
not modelled
65.1
11
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Allosteric threonine deaminase C-terminal domain81 d2hmfa3
not modelled
64.9
19
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Aspartokinase allosteric domain-like82 c3c1nA_
not modelled
64.5
17
PDB header: transferaseChain: A: PDB Molecule: probable aspartokinase;PDBTitle: crystal structure of allosteric inhibition threonine-sensitive2 aspartokinase from methanococcus jannaschii with l-threonine
83 c2re1A_
not modelled
63.7
10
PDB header: transferaseChain: A: PDB Molecule: aspartokinase, alpha and beta subunits;PDBTitle: crystal structure of aspartokinase alpha and beta subunits
84 c2v25B_
not modelled
61.7
11
PDB header: receptorChain: B: PDB Molecule: major cell-binding factor;PDBTitle: structure of the campylobacter jejuni antigen peb1a, an2 aspartate and glutamate receptor with bound aspartate
85 c3m05A_
not modelled
57.9
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein pepe_1480;PDBTitle: the crystal structure of a functionally unknown protein2 pepe_1480 from pediococcus pentosaceus atcc 25745
86 d2cdqa2
not modelled
56.7
18
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Aspartokinase allosteric domain-like87 c2x7pA_
not modelled
54.4
14
PDB header: unknown functionChain: A: PDB Molecule: possible thiamine biosynthesis enzyme;PDBTitle: the conserved candida albicans ca3427 gene product defines a new2 family of proteins exhibiting the generic periplasmic binding3 protein structural fold
88 c2cdqB_
not modelled
52.1
14
PDB header: transferaseChain: B: PDB Molecule: aspartokinase;PDBTitle: crystal structure of arabidopsis thaliana aspartate kinase2 complexed with lysine and s-adenosylmethionine
89 c1j8yF_
not modelled
45.4
12
PDB header: signaling proteinChain: F: PDB Molecule: signal recognition 54 kda protein;PDBTitle: signal recognition particle conserved gtpase domain from a.2 ambivalens t112a mutant
90 d2hmfa2
not modelled
45.2
9
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Aspartokinase allosteric domain-like91 c2xu6B_
not modelled
43.8
10
PDB header: protein bindingChain: B: PDB Molecule: mdv1 coiled coil;PDBTitle: mdv1 coiled coil domain
92 c3mahA_
not modelled
42.4
6
PDB header: transferaseChain: A: PDB Molecule: aspartokinase;PDBTitle: a putative c-terminal regulatory domain of aspartate kinase from2 porphyromonas gingivalis w83.
93 c3kzgB_
not modelled
42.4
13
PDB header: transport proteinChain: B: PDB Molecule: arginine 3rd transport system periplasmic bindingPDBTitle: crystal structure of an arginine 3rd transport system2 periplasmic binding protein from legionella pneumophila
94 c2yhsA_
not modelled
41.0
16
PDB header: cell cycleChain: A: PDB Molecule: cell division protein ftsy;PDBTitle: structure of the e. coli srp receptor ftsy
95 d2a5sa1
not modelled
38.8
11
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like96 d2j0wa2
not modelled
38.7
23
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Aspartokinase allosteric domain-like97 c2og2A_
not modelled
38.5
15
PDB header: protein transportChain: A: PDB Molecule: putative signal recognition particle receptor;PDBTitle: crystal structure of chloroplast ftsy from arabidopsis2 thaliana
98 c2i4cA_
not modelled
36.9
13
PDB header: transport proteinChain: A: PDB Molecule: bicarbonate transporter;PDBTitle: crystal structure of bicarbonate transport protein cmpa from2 synechocystis sp. pcc 6803 in complex with bicarbonate and calcium
99 c2ke4A_
not modelled
35.4
10
PDB header: membrane proteinChain: A: PDB Molecule: cdc42-interacting protein 4;PDBTitle: the nmr structure of the tc10 and cdc42 interacting domain2 of cip4
100 c3h1gA_
not modelled
34.2
13
PDB header: signaling proteinChain: A: PDB Molecule: chemotaxis protein chey homolog;PDBTitle: crystal structure of chey mutant t84a of helicobacter pylori
101 c3r84O_
not modelled
33.7
16
PDB header: transcriptionChain: O: PDB Molecule: mediator of rna polymerase ii transcription subunit 11;PDBTitle: structure of the mediator head subcomplex med11/22
102 c2rc9A_
not modelled
30.8
11
PDB header: membrane proteinChain: A: PDB Molecule: glutamate [nmda] receptor subunit 3a;PDBTitle: crystal structure of the nr3a ligand binding core complex with acpc at2 1.96 angstrom resolution
103 d2czla1
not modelled
27.4
9
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like104 c3k5pA_
not modelled
25.4
16
PDB header: oxidoreductaseChain: A: PDB Molecule: d-3-phosphoglycerate dehydrogenase;PDBTitle: crystal structure of amino acid-binding act: d-isomer specific 2-2 hydroxyacid dehydrogenase catalytic domain from brucella melitensis
105 d1ozja_
not modelled
25.4
10
Fold: SMAD MH1 domainSuperfamily: SMAD MH1 domainFamily: SMAD MH1 domain106 c2cnwF_
not modelled
25.4
11
PDB header: signal recognitionChain: F: PDB Molecule: cell division protein ftsy;PDBTitle: gdpalf4 complex of the srp gtpases ffh and ftsy
107 c3i42A_
not modelled
24.9
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: response regulator receiver domain protein (chey-PDBTitle: structure of response regulator receiver domain (chey-like)2 from methylobacillus flagellatus
108 d3dhxa1
not modelled
23.9
23
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: NIL domain-like109 c2rejA_
not modelled
23.6
11
PDB header: choline-binding proteinChain: A: PDB Molecule: putative glycine betaine abc transporter protein;PDBTitle: abc-transporter choline binding protein in unliganded semi-2 closed conformation
110 c2wmyH_
not modelled
23.4
16
PDB header: hydrolaseChain: H: PDB Molecule: putative acid phosphatase wzb;PDBTitle: crystal structure of the tyrosine phosphatase wzb from2 escherichia coli k30 in complex with sulphate.
111 d1qnta2
not modelled
22.6
50
Fold: Ribonuclease H-like motifSuperfamily: Methylated DNA-protein cysteine methyltransferase domainFamily: Methylated DNA-protein cysteine methyltransferase domain112 d1w36d1
not modelled
21.9
16
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain113 d1nwpa_
not modelled
21.5
6
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Plastocyanin/azurin-like114 d2qswa1
not modelled
21.3
16
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: NIL domain-like115 c1gk4A_
not modelled
21.3
14
PDB header: vimentinChain: A: PDB Molecule: vimentin;PDBTitle: human vimentin coil 2b fragment (cys2)
116 c2ia0A_
not modelled
21.2
33
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative hth-type transcriptional regulator pf0864;PDBTitle: transcriptional regulatory protein pf0864 from pyrococcus furiosus a2 member of the asnc family (pf0864)
117 d1us5a_
not modelled
21.0
15
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like118 c3hv1A_
not modelled
20.6
12
PDB header: transport proteinChain: A: PDB Molecule: polar amino acid abc uptake transporter substratePDBTitle: crystal structure of a polar amino acid abc uptake2 transporter substrate binding protein from streptococcus3 thermophilus