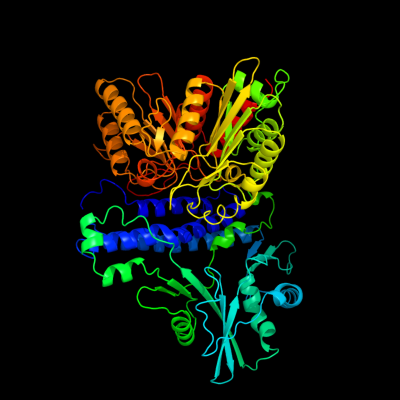
| 1 | c1xdoB_
|
|
 |
100.0 |
100 |
PDB header:transferase
Chain: B: PDB Molecule:polyphosphate kinase;
PDBTitle: crystal structure of escherichia coli polyphosphate kinase
|
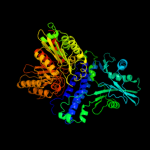
| 2 | c2o8rA_
|
|
 |
100.0 |
38 |
PDB header:transferase
Chain: A: PDB Molecule:polyphosphate kinase;
PDBTitle: crystal structure of polyphosphate kinase from2 porphyromonas gingivalis
|
| 3 | d1xdpa4
|
|
 |
100.0 |
100 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Polyphosphate kinase C-terminal domain |
| 4 | d2o8ra4
|
|
 |
100.0 |
43 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Polyphosphate kinase C-terminal domain |
| 5 | d2o8ra3
|
|
 |
100.0 |
39 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Polyphosphate kinase C-terminal domain |
| 6 | d1xdpa2
|
|
 |
100.0 |
100 |
Fold:PHP14-like
Superfamily:PHP14-like
Family:PPK middle domain-like |
| 7 | d1xdpa3
|
|
 |
100.0 |
100 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Polyphosphate kinase C-terminal domain |
| 8 | d2o8ra2
|
|
 |
100.0 |
30 |
Fold:PHP14-like
Superfamily:PHP14-like
Family:PPK middle domain-like |
| 9 | d1xdpa1
|
|
 |
100.0 |
100 |
Fold:Spectrin repeat-like
Superfamily:PPK N-terminal domain-like
Family:PPK N-terminal domain-like |
| 10 | d2o8ra1
|
|
 |
100.0 |
38 |
Fold:Spectrin repeat-like
Superfamily:PPK N-terminal domain-like
Family:PPK N-terminal domain-like |
| 11 | c3hsiC_
|
|
 |
99.9 |
17 |
PDB header:transferase
Chain: C: PDB Molecule:phosphatidylserine synthase;
PDBTitle: crystal structure of phosphatidylserine synthase haemophilus2 influenzae rd kw20
|
| 12 | c1v0sA_
|
|
 |
99.9 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:phospholipase d;
PDBTitle: uninhibited form of phospholipase d from streptomyces sp.2 strain pmf
|
| 13 | d1byra_
|
|
 |
99.4 |
15 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Nuclease |
| 14 | d1v0wa2
|
|
 |
99.1 |
17 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Phospholipase D |
| 15 | d1v0wa1
|
|
 |
99.0 |
14 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Phospholipase D |
| 16 | c2c1lA_
|
|
 |
92.3 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:restriction endonuclease;
PDBTitle: structure of the bfii restriction endonuclease
|
| 17 | c2yv2A_
|
|
 |
91.5 |
21 |
PDB header:ligase
Chain: A: PDB Molecule:succinyl-coa synthetase alpha chain;
PDBTitle: crystal structure of succinyl-coa synthetase alpha chain from2 aeropyrum pernix k1
|
| 18 | d1vb5a_
|
|
 |
91.0 |
19 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:IF2B-like |
| 19 | c3ecsD_
|
|
 |
89.5 |
25 |
PDB header:translation
Chain: D: PDB Molecule:translation initiation factor eif-2b subunit
PDBTitle: crystal structure of human eif2b alpha
|
| 20 | d2csua2
|
|
 |
87.9 |
15 |
Fold:Flavodoxin-like
Superfamily:Succinyl-CoA synthetase domains
Family:Succinyl-CoA synthetase domains |
| 21 | d1eucb1 |
|
not modelled |
87.7 |
29 |
Fold:Flavodoxin-like
Superfamily:Succinyl-CoA synthetase domains
Family:Succinyl-CoA synthetase domains |
| 22 | c2fpgA_ |
|
not modelled |
86.9 |
20 |
PDB header:ligase
Chain: A: PDB Molecule:succinyl-coa ligase [gdp-forming] alpha-chain,
PDBTitle: crystal structure of pig gtp-specific succinyl-coa2 synthetase in complex with gdp
|
| 23 | c3a11D_ |
|
not modelled |
86.2 |
28 |
PDB header:isomerase
Chain: D: PDB Molecule:translation initiation factor eif-2b, delta
PDBTitle: crystal structure of ribose-1,5-bisphosphate isomerase from2 thermococcus kodakaraensis kod1
|
| 24 | d1oi7a2 |
|
not modelled |
85.9 |
16 |
Fold:Flavodoxin-like
Superfamily:Succinyl-CoA synthetase domains
Family:Succinyl-CoA synthetase domains |
| 25 | d1t9ka_ |
|
not modelled |
85.7 |
18 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:IF2B-like |
| 26 | c3r2jC_ |
|
not modelled |
84.8 |
27 |
PDB header:hydrolase
Chain: C: PDB Molecule:alpha/beta-hydrolase-like protein;
PDBTitle: crystal structure of pnc1 from l. infantum in complex with nicotinate
|
| 27 | d2nu7a2 |
|
not modelled |
84.7 |
18 |
Fold:Flavodoxin-like
Superfamily:Succinyl-CoA synthetase domains
Family:Succinyl-CoA synthetase domains |
| 28 | c2yboA_ |
|
not modelled |
84.7 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:methyltransferase;
PDBTitle: the x-ray structure of the sam-dependent uroporphyrinogen2 iii methyltransferase nire from pseudomonas aeruginosa in3 complex with sah
|
| 29 | d1zj8a2 |
|
not modelled |
84.6 |
20 |
Fold:Ferredoxin-like
Superfamily:Nitrite/Sulfite reductase N-terminal domain-like
Family:Duplicated SiR/NiR-like domains 1 and 3 |
| 30 | d1t5oa_ |
|
not modelled |
84.5 |
18 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:IF2B-like |
| 31 | d2a0ua1 |
|
not modelled |
82.9 |
8 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:IF2B-like |
| 32 | c2nu8D_ |
|
not modelled |
82.8 |
16 |
PDB header:ligase
Chain: D: PDB Molecule:succinyl-coa ligase [adp-forming] subunit alpha;
PDBTitle: c123at mutant of e. coli succinyl-coa synthetase
|
| 33 | c3mwdB_ |
|
not modelled |
81.4 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:atp-citrate synthase;
PDBTitle: truncated human atp-citrate lyase with citrate bound
|
| 34 | c2yvkA_ |
|
not modelled |
81.3 |
12 |
PDB header:isomerase
Chain: A: PDB Molecule:methylthioribose-1-phosphate isomerase;
PDBTitle: crystal structure of 5-methylthioribose 1-phosphate2 isomerase product complex from bacillus subtilis
|
| 35 | c2csuB_ |
|
not modelled |
80.8 |
15 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:457aa long hypothetical protein;
PDBTitle: crystal structure of ph0766 from pyrococcus horikoshii ot3
|
| 36 | c1yd6A_ |
|
not modelled |
80.3 |
18 |
PDB header:dna binding protein
Chain: A: PDB Molecule:uvrc;
PDBTitle: crystal structure of the giy-yig n-terminal endonuclease2 domain of uvrc from bacillus caldotenax
|
| 37 | c3gbcA_ |
|
not modelled |
78.9 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:pyrazinamidase/nicotinamidas pnca;
PDBTitle: determination of the crystal structure of the pyrazinamidase from2 m.tuberculosis : a structure-function analysis for prediction3 resistance to pyrazinamide
|
| 38 | c1oi7A_ |
|
not modelled |
78.8 |
16 |
PDB header:synthetase
Chain: A: PDB Molecule:succinyl-coa synthetase alpha chain;
PDBTitle: the crystal structure of succinyl-coa synthetase alpha2 subunit from thermus thermophilus
|
| 39 | d1s4da_ |
|
not modelled |
78.8 |
17 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
| 40 | c1yd2A_ |
|
not modelled |
77.8 |
33 |
PDB header:dna binding protein
Chain: A: PDB Molecule:uvrabc system protein c;
PDBTitle: crystal structure of the giy-yig n-terminal endonuclease domain of2 uvrc from thermotoga maritima: point mutant y19f bound to the3 catalytic divalent cation
|
| 41 | d2nu7b1 |
|
not modelled |
75.6 |
30 |
Fold:Flavodoxin-like
Superfamily:Succinyl-CoA synthetase domains
Family:Succinyl-CoA synthetase domains |
| 42 | c2b34C_ |
|
not modelled |
75.5 |
14 |
PDB header:hydrolase
Chain: C: PDB Molecule:mar1 ribonuclease;
PDBTitle: structure of mar1 ribonuclease from caenorhabditis elegans
|
| 43 | c3ndcB_ |
|
not modelled |
73.4 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:precorrin-4 c(11)-methyltransferase;
PDBTitle: crystal structure of precorrin-4 c11-methyltransferase from2 rhodobacter capsulatus
|
| 44 | c1nvmG_ |
|
not modelled |
73.3 |
9 |
PDB header:lyase/oxidoreductase
Chain: G: PDB Molecule:4-hydroxy-2-oxovalerate aldolase;
PDBTitle: crystal structure of a bifunctional aldolase-dehydrogenase :2 sequestering a reactive and volatile intermediate
|
| 45 | c1zj8B_ |
|
not modelled |
68.7 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable ferredoxin-dependent nitrite reductase nira;
PDBTitle: structure of mycobacterium tuberculosis nira protein
|
| 46 | c1eucB_ |
|
not modelled |
68.5 |
28 |
PDB header:ligase
Chain: B: PDB Molecule:succinyl-coa synthetase, beta chain;
PDBTitle: crystal structure of dephosphorylated pig heart, gtp-2 specific succinyl-coa synthetase
|
| 47 | d1jy1a1 |
|
not modelled |
67.4 |
13 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Tyrosyl-DNA phosphodiesterase TDP1 |
| 48 | d1qzqa1 |
|
not modelled |
67.1 |
13 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Tyrosyl-DNA phosphodiesterase TDP1 |
| 49 | d1euca2 |
|
not modelled |
65.1 |
20 |
Fold:Flavodoxin-like
Superfamily:Succinyl-CoA synthetase domains
Family:Succinyl-CoA synthetase domains |
| 50 | d2akja2 |
|
not modelled |
63.1 |
17 |
Fold:Ferredoxin-like
Superfamily:Nitrite/Sulfite reductase N-terminal domain-like
Family:Duplicated SiR/NiR-like domains 1 and 3 |
| 51 | c1yzvA_ |
|
not modelled |
63.0 |
11 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein;
PDBTitle: hypothetical protein from trypanosoma cruzi
|
| 52 | d1vhna_ |
|
not modelled |
62.3 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 53 | d1jpma1 |
|
not modelled |
60.3 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
| 54 | d1nvma2 |
|
not modelled |
56.9 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:HMGL-like |
| 55 | d2nu7a1 |
|
not modelled |
56.3 |
24 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
| 56 | c2h0rD_ |
|
not modelled |
55.9 |
19 |
PDB header:hydrolase
Chain: D: PDB Molecule:nicotinamidase;
PDBTitle: structure of the yeast nicotinamidase pnc1p
|
| 57 | c2rqmA_ |
|
not modelled |
54.7 |
24 |
PDB header:chaperone
Chain: A: PDB Molecule:mesoderm development candidate 2;
PDBTitle: nmr solution structure of mesoderm development (mesd) - open2 conformation
|
| 58 | d1x9ga_ |
|
not modelled |
53.7 |
9 |
Fold:Isochorismatase-like hydrolases
Superfamily:Isochorismatase-like hydrolases
Family:Isochorismatase-like hydrolases |
| 59 | d1yaca_ |
|
not modelled |
52.2 |
13 |
Fold:Isochorismatase-like hydrolases
Superfamily:Isochorismatase-like hydrolases
Family:Isochorismatase-like hydrolases |
| 60 | c3d3kD_ |
|
not modelled |
51.1 |
12 |
PDB header:protein binding
Chain: D: PDB Molecule:enhancer of mrna-decapping protein 3;
PDBTitle: crystal structure of human edc3p
|
| 61 | d1ebda2 |
|
not modelled |
50.7 |
30 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 62 | c3kwpA_ |
|
not modelled |
50.4 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:predicted methyltransferase;
PDBTitle: crystal structure of putative methyltransferase from lactobacillus2 brevis
|
| 63 | d1im5a_ |
|
not modelled |
50.1 |
17 |
Fold:Isochorismatase-like hydrolases
Superfamily:Isochorismatase-like hydrolases
Family:Isochorismatase-like hydrolases |
| 64 | d1ve2a1 |
|
not modelled |
48.8 |
15 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
| 65 | c2hxtA_ |
|
not modelled |
47.7 |
25 |
PDB header:unknown function
Chain: A: PDB Molecule:l-fuconate dehydratase;
PDBTitle: crystal structure of l-fuconate dehydratase from xanthomonas2 campestris liganded with mg++ and d-erythronohydroxamate
|
| 66 | c2pjuD_ |
|
not modelled |
47.5 |
19 |
PDB header:transcription
Chain: D: PDB Molecule:propionate catabolism operon regulatory protein;
PDBTitle: crystal structure of propionate catabolism operon2 regulatory protein prpr
|
| 67 | c1rr2A_ |
|
not modelled |
46.7 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:transcarboxylase 5s subunit;
PDBTitle: propionibacterium shermanii transcarboxylase 5s subunit bound to 2-2 ketobutyric acid
|
| 68 | c2nu9E_ |
|
not modelled |
46.2 |
34 |
PDB header:ligase
Chain: E: PDB Molecule:succinyl-coa synthetase beta chain;
PDBTitle: c123at mutant of e. coli succinyl-coa synthetase2 orthorhombic crystal form
|
| 69 | d1va0a1 |
|
not modelled |
45.8 |
17 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
| 70 | c2q5cA_ |
|
not modelled |
45.7 |
13 |
PDB header:transcription
Chain: A: PDB Molecule:ntrc family transcriptional regulator;
PDBTitle: crystal structure of ntrc family transcriptional regulator from2 clostridium acetobutylicum
|
| 71 | d1dxla2 |
|
not modelled |
45.1 |
20 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 72 | d1aoga2 |
|
not modelled |
44.4 |
19 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 73 | c2wtaA_ |
|
not modelled |
44.1 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:nicotinamidase;
PDBTitle: acinetobacter baumanii nicotinamidase pyrazinamidease
|
| 74 | d1lvla2 |
|
not modelled |
43.5 |
18 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 75 | d1mk0a_ |
|
not modelled |
41.4 |
32 |
Fold:GIY-YIG endonuclease
Superfamily:GIY-YIG endonuclease
Family:GIY-YIG endonuclease |
| 76 | c3dmyA_ |
|
not modelled |
41.2 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:protein fdra;
PDBTitle: crystal structure of a predicated acyl-coa synthetase from e.coli
|
| 77 | d1v59a2 |
|
not modelled |
41.1 |
25 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 78 | c1cbfA_ |
|
not modelled |
40.8 |
17 |
PDB header:methyltransferase
Chain: A: PDB Molecule:cobalt-precorrin-4 transmethylase;
PDBTitle: the x-ray structure of a cobalamin biosynthetic enzyme, cobalt2 precorrin-4 methyltransferase, cbif
|
| 79 | d1cbfa_ |
|
not modelled |
40.8 |
17 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
| 80 | c1nopB_ |
|
not modelled |
39.2 |
13 |
PDB header:hydrolase/dna
Chain: B: PDB Molecule:tyrosyl-dna phosphodiesterase 1;
PDBTitle: crystal structure of human tyrosyl-dna phosphodiesterase2 (tdp1) in complex with vanadate, dna and a human3 topoisomerase i-derived peptide
|
| 81 | c3eegB_ |
|
not modelled |
38.1 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:2-isopropylmalate synthase;
PDBTitle: crystal structure of a 2-isopropylmalate synthase from2 cytophaga hutchinsonii
|
| 82 | c2jlaD_ |
|
not modelled |
37.8 |
57 |
PDB header:transferase
Chain: D: PDB Molecule:2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene
PDBTitle: crystal structure of e.coli mend, 2-succinyl-5-enolpyruvyl-2 6-hydroxy-3-cyclohexadiene-1-carboxylate synthase - semet3 protein
|
| 83 | c1nu5A_ |
|
not modelled |
37.7 |
29 |
PDB header:isomerase
Chain: A: PDB Molecule:chloromuconate cycloisomerase;
PDBTitle: crystal structure of pseudomonas sp. p51 chloromuconate lactonizing2 enzyme
|
| 84 | d2pjua1 |
|
not modelled |
36.9 |
19 |
Fold:Chelatase-like
Superfamily:PrpR receptor domain-like
Family:PrpR receptor domain-like |
| 85 | d1oi7a1 |
|
not modelled |
36.8 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
| 86 | d1djqa2 |
|
not modelled |
35.7 |
20 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:C-terminal domain of adrenodoxin reductase-like |
| 87 | c2v4jA_ |
|
not modelled |
35.5 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:sulfite reductase, dissimilatory-type subunit
PDBTitle: the crystal structure of desulfovibrio vulgaris2 dissimilatory sulfite reductase bound to dsrc provides3 novel insights into the mechanism of sulfate respiration
|
| 88 | c2akjA_ |
|
not modelled |
35.2 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ferredoxin--nitrite reductase, chloroplast;
PDBTitle: structure of spinach nitrite reductase
|
| 89 | d1oy0a_ |
|
not modelled |
35.2 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Ketopantoate hydroxymethyltransferase PanB |
| 90 | d3lada2 |
|
not modelled |
33.4 |
21 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 91 | d2d59a1 |
|
not modelled |
31.7 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
| 92 | d1ojta2 |
|
not modelled |
31.2 |
19 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 93 | c2qvhB_ |
|
not modelled |
30.6 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:o-succinylbenzoate-coa synthase;
PDBTitle: crystal structure of o-succinylbenzoate synthase complexed with o-2 succinyl benzoate (osb)
|
| 94 | d1j2ra_ |
|
not modelled |
30.4 |
9 |
Fold:Isochorismatase-like hydrolases
Superfamily:Isochorismatase-like hydrolases
Family:Isochorismatase-like hydrolases |
| 95 | c3msyC_ |
|
not modelled |
30.3 |
11 |
PDB header:isomerase
Chain: C: PDB Molecule:mandelate racemase/muconate lactonizing enzyme;
PDBTitle: crystal structure of mandelate racemase/muconate lactonizing enzyme2 from a marine actinobacterium
|
| 96 | c3ivuB_ |
|
not modelled |
30.1 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:homocitrate synthase, mitochondrial;
PDBTitle: homocitrate synthase lys4 bound to 2-og
|
| 97 | c2kglA_ |
|
not modelled |
29.9 |
24 |
PDB header:chaperone
Chain: A: PDB Molecule:mesoderm development candidate 2;
PDBTitle: nmr solution structure of mesd
|
| 98 | c3ot4F_ |
|
not modelled |
28.7 |
9 |
PDB header:hydrolase
Chain: F: PDB Molecule:putative isochorismatase;
PDBTitle: structure and catalytic mechanism of bordetella bronchiseptica nicf
|
| 99 | d1pjqa2 |
|
not modelled |
28.5 |
16 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
| 100 | c2i9sA_ |
|
not modelled |
27.5 |
22 |
PDB header:chaperone
Chain: A: PDB Molecule:mesoderm development candidate 2;
PDBTitle: the solution structure of the core of mesoderm development2 (mesd).
|
| 101 | c1sy7B_ |
|
not modelled |
27.1 |
9 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:catalase 1;
PDBTitle: crystal structure of the catalase-1 from neurospora crassa, native2 structure at 1.75a resolution.
|
| 102 | c2dc1A_ |
|
not modelled |
27.1 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-aspartate dehydrogenase;
PDBTitle: crystal structure of l-aspartate dehydrogenase from2 hyperthermophilic archaeon archaeoglobus fulgidus
|
| 103 | d1lpfa2 |
|
not modelled |
26.2 |
24 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 104 | d1d7ya2 |
|
not modelled |
25.8 |
21 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 105 | c1vixA_ |
|
not modelled |
25.4 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptidase t;
PDBTitle: crystal structure of a putative peptidase t
|
| 106 | d3grsa2 |
|
not modelled |
25.2 |
17 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 107 | d1r6wa1 |
|
not modelled |
25.2 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
| 108 | c2yv1A_ |
|
not modelled |
25.1 |
16 |
PDB header:ligase
Chain: A: PDB Molecule:succinyl-coa ligase [adp-forming] subunit alpha;
PDBTitle: crystal structure of succinyl-coa synthetase alpha chain from2 methanocaldococcus jannaschii dsm 2661
|
| 109 | d1zpda2 |
|
not modelled |
24.6 |
35 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Pyruvate oxidase and decarboxylase Pyr module |
| 110 | d1t9ba2 |
|
not modelled |
24.4 |
24 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Pyruvate oxidase and decarboxylase Pyr module |
| 111 | c3bezC_ |
|
not modelled |
24.4 |
27 |
PDB header:hydrolase
Chain: C: PDB Molecule:protease 4;
PDBTitle: crystal structure of escherichia coli signal peptide peptidase (sppa),2 semet crystals
|
| 112 | c1pjtB_ |
|
not modelled |
23.9 |
16 |
PDB header:transferase/oxidoreductase/lyase
Chain: B: PDB Molecule:siroheme synthase;
PDBTitle: the structure of the ser128ala point-mutant variant of cysg,2 the multifunctional3 methyltransferase/dehydrogenase/ferrochelatase for4 siroheme synthesis
|
| 113 | c2frxD_ |
|
not modelled |
23.8 |
5 |
PDB header:transferase
Chain: D: PDB Molecule:hypothetical protein yebu;
PDBTitle: crystal structure of yebu, a m5c rna methyltransferase from e.coli
|
| 114 | c3ewbX_ |
|
not modelled |
23.3 |
23 |
PDB header:transferase
Chain: X: PDB Molecule:2-isopropylmalate synthase;
PDBTitle: crystal structure of n-terminal domain of putative 2-2 isopropylmalate synthase from listeria monocytogenes
|
| 115 | d1onfa2 |
|
not modelled |
23.2 |
9 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 116 | d1xhca2 |
|
not modelled |
22.6 |
19 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 117 | d1tuea_ |
|
not modelled |
22.5 |
27 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Extended AAA-ATPase domain |
| 118 | d1tzza1 |
|
not modelled |
22.5 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
| 119 | d2ax3a2 |
|
not modelled |
22.5 |
21 |
Fold:YjeF N-terminal domain-like
Superfamily:YjeF N-terminal domain-like
Family:YjeF N-terminal domain-like |
| 120 | d3c7bb2 |
|
not modelled |
22.3 |
24 |
Fold:Ferredoxin-like
Superfamily:Nitrite/Sulfite reductase N-terminal domain-like
Family:DsrA/DsrB N-terminal-domain-like |