| 1 | c2ashB_
|
|
 |
100.0 |
49 |
PDB header:transferase
Chain: B: PDB Molecule:queuine trna-ribosyltransferase;
PDBTitle: crystal structure of queuine trna-ribosyltransferase (ec 2.4.2.29)2 (trna-guanine (tm1561) from thermotoga maritima at 1.90 a resolution
|
| 2 | d1r5ya_
|
|
 |
100.0 |
58 |
Fold:TIM beta/alpha-barrel
Superfamily:tRNA-guanine transglycosylase
Family:tRNA-guanine transglycosylase |
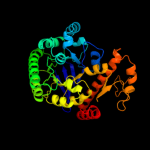
| 3 | c1iq8B_
|
|
 |
100.0 |
28 |
PDB header:transferase
Chain: B: PDB Molecule:archaeosine trna-guanine transglycosylase;
PDBTitle: crystal structure of archaeosine trna-guanine2 transglycosylase from pyrococcus horikoshii
|
| 4 | d1iq8a1
|
|
 |
100.0 |
29 |
Fold:TIM beta/alpha-barrel
Superfamily:tRNA-guanine transglycosylase
Family:tRNA-guanine transglycosylase |
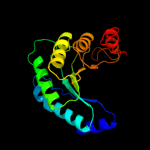
| 5 | c3bg3B_
|
|
 |
95.9 |
14 |
PDB header:ligase
Chain: B: PDB Molecule:pyruvate carboxylase, mitochondrial;
PDBTitle: crystal structure of human pyruvate carboxylase (missing2 the biotin carboxylase domain at the n-terminus)
|
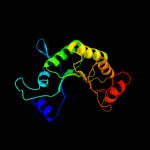
| 6 | c1rr2A_
|
|
 |
95.2 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:transcarboxylase 5s subunit;
PDBTitle: propionibacterium shermanii transcarboxylase 5s subunit bound to 2-2 ketobutyric acid
|
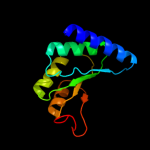
| 7 | c3ewbX_
|
|
 |
94.4 |
10 |
PDB header:transferase
Chain: X: PDB Molecule:2-isopropylmalate synthase;
PDBTitle: crystal structure of n-terminal domain of putative 2-2 isopropylmalate synthase from listeria monocytogenes
|
| 8 | c1ydnA_
|
|
 |
93.3 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:hydroxymethylglutaryl-coa lyase;
PDBTitle: crystal structure of the hmg-coa lyase from brucella melitensis,2 northeast structural genomics target lr35.
|
| 9 | c1ydoC_
|
|
 |
92.3 |
13 |
PDB header:lyase
Chain: C: PDB Molecule:hmg-coa lyase;
PDBTitle: crystal structure of the bacillis subtilis hmg-coa lyase, northeast2 structural genomics target sr181.
|
| 10 | c3fluD_
|
|
 |
91.8 |
12 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from the pathogen2 neisseria meningitidis
|
| 11 | c3eegB_
|
|
 |
91.7 |
10 |
PDB header:transferase
Chain: B: PDB Molecule:2-isopropylmalate synthase;
PDBTitle: crystal structure of a 2-isopropylmalate synthase from2 cytophaga hutchinsonii
|
| 12 | c2ftpA_
|
|
 |
91.4 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:hydroxymethylglutaryl-coa lyase;
PDBTitle: crystal structure of hydroxymethylglutaryl-coa lyase from pseudomonas2 aeruginosa
|
| 13 | d1qapa1
|
|
 |
90.7 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Nicotinate/Quinolinate PRTase C-terminal domain-like
Family:NadC C-terminal domain-like |
| 14 | d1sr9a2
|
|
 |
90.5 |
7 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:HMGL-like |
| 15 | c3navB_
|
|
 |
90.4 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of an alpha subunit of tryptophan synthase from2 vibrio cholerae o1 biovar el tor str. n16961
|
| 16 | c1nvmG_
|
|
 |
90.0 |
14 |
PDB header:lyase/oxidoreductase
Chain: G: PDB Molecule:4-hydroxy-2-oxovalerate aldolase;
PDBTitle: crystal structure of a bifunctional aldolase-dehydrogenase :2 sequestering a reactive and volatile intermediate
|
| 17 | c3bg5C_
|
|
 |
89.3 |
17 |
PDB header:ligase
Chain: C: PDB Molecule:pyruvate carboxylase;
PDBTitle: crystal structure of staphylococcus aureus pyruvate2 carboxylase
|
| 18 | d1nvma2
|
|
 |
88.5 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:HMGL-like |
| 19 | c2cw6B_
|
|
 |
86.9 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:hydroxymethylglutaryl-coa lyase, mitochondrial;
PDBTitle: crystal structure of human hmg-coa lyase: insights into2 catalysis and the molecular basis for3 hydroxymethylglutaric aciduria
|
| 20 | c3pueA_
|
|
 |
86.7 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of the complex of dhydrodipicolinate synthase from2 acinetobacter baumannii with lysine at 2.6a resolution
|
| 21 | c3l0gD_ |
|
not modelled |
86.3 |
9 |
PDB header:transferase
Chain: D: PDB Molecule:nicotinate-nucleotide pyrophosphorylase;
PDBTitle: crystal structure of nicotinate-nucleotide pyrophosphorylase from2 ehrlichia chaffeensis at 2.05a resolution
|
| 22 | d1goxa_ |
|
not modelled |
86.3 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 23 | c2ehhE_ |
|
not modelled |
85.8 |
15 |
PDB header:lyase
Chain: E: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 aquifex aeolicus
|
| 24 | c3qjaA_ |
|
not modelled |
85.4 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:indole-3-glycerol phosphate synthase;
PDBTitle: crystal structure of the mycobacterium tuberculosis indole-3-glycerol2 phosphate synthase (trpc) in apo form
|
| 25 | d1d3ga_ |
|
not modelled |
85.3 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 26 | c3na8A_ |
|
not modelled |
84.7 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:putative dihydrodipicolinate synthetase;
PDBTitle: crystal structure of a putative dihydrodipicolinate synthetase from2 pseudomonas aeruginosa
|
| 27 | c1qapA_ |
|
not modelled |
84.4 |
14 |
PDB header:glycosyltransferase
Chain: A: PDB Molecule:quinolinic acid phosphoribosyltransferase;
PDBTitle: quinolinic acid phosphoribosyltransferase with bound2 quinolinic acid
|
| 28 | d1uuma_ |
|
not modelled |
84.1 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 29 | c3bleA_ |
|
not modelled |
83.4 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:citramalate synthase from leptospira interrogans;
PDBTitle: crystal structure of the catalytic domain of licms in2 complexed with malonate
|
| 30 | d1tb3a1 |
|
not modelled |
83.0 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 31 | d1geqa_ |
|
not modelled |
82.9 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 32 | d1a53a_ |
|
not modelled |
82.0 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 33 | d1xxxa1 |
|
not modelled |
81.8 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 34 | c2cdh1_ |
|
not modelled |
81.1 |
20 |
PDB header:transferase
Chain: 1: PDB Molecule:enoyl reductase;
PDBTitle: architecture of the thermomyces lanuginosus fungal fatty2 acid synthase at 5 angstrom resolution.
|
| 35 | d1o4ua1 |
|
not modelled |
80.7 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Nicotinate/Quinolinate PRTase C-terminal domain-like
Family:NadC C-terminal domain-like |
| 36 | c2vc6A_ |
|
not modelled |
80.5 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: structure of mosa from s. meliloti with pyruvate bound
|
| 37 | c1x1oC_ |
|
not modelled |
80.4 |
14 |
PDB header:transferase
Chain: C: PDB Molecule:nicotinate-nucleotide pyrophosphorylase;
PDBTitle: crystal structure of project id tt0268 from thermus thermophilus hb8
|
| 38 | c3cprB_ |
|
not modelled |
80.3 |
18 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthetase;
PDBTitle: the crystal structure of corynebacterium glutamicum2 dihydrodipicolinate synthase to 2.2 a resolution
|
| 39 | d1o5ka_ |
|
not modelled |
80.2 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 40 | c2zyfA_ |
|
not modelled |
80.1 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:homocitrate synthase;
PDBTitle: crystal structure of homocitrate synthase from thermus thermophilus2 complexed with magnesuim ion and alpha-ketoglutarate
|
| 41 | c3tqvA_ |
|
not modelled |
80.1 |
9 |
PDB header:transferase
Chain: A: PDB Molecule:nicotinate-nucleotide pyrophosphorylase;
PDBTitle: structure of the nicotinate-nucleotide pyrophosphorylase from2 francisella tularensis.
|
| 42 | c2e77B_ |
|
not modelled |
79.4 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:lactate oxidase;
PDBTitle: crystal structure of l-lactate oxidase with pyruvate complex
|
| 43 | c3hf3A_ |
|
not modelled |
79.3 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:chromate reductase;
PDBTitle: old yellow enzyme from thermus scotoductus sa-01
|
| 44 | d1qpoa1 |
|
not modelled |
78.9 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Nicotinate/Quinolinate PRTase C-terminal domain-like
Family:NadC C-terminal domain-like |
| 45 | d1rqba2 |
|
not modelled |
78.8 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:HMGL-like |
| 46 | c3si9B_ |
|
not modelled |
78.3 |
22 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from bartonella2 henselae
|
| 47 | c3mcnA_ |
|
not modelled |
78.1 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:2-amino-4-hydroxy-6-hydroxymethyldihydropteridine
PDBTitle: crystal structure of the 6-hyroxymethyl-7,8-dihydropterin2 pyrophosphokinase dihydropteroate synthase bifunctional enzyme from3 francisella tularensis
|
| 48 | c2rduA_ |
|
not modelled |
78.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:hydroxyacid oxidase 1;
PDBTitle: crystal structure of human glycolate oxidase in complex with2 glyoxylate
|
| 49 | c3lciA_ |
|
not modelled |
77.8 |
22 |
PDB header:lyase
Chain: A: PDB Molecule:n-acetylneuraminate lyase;
PDBTitle: the d-sialic acid aldolase mutant v251w
|
| 50 | c3pajA_ |
|
not modelled |
77.5 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:nicotinate-nucleotide pyrophosphorylase, carboxylating;
PDBTitle: 2.00 angstrom resolution crystal structure of a quinolinate2 phosphoribosyltransferase from vibrio cholerae o1 biovar eltor str.3 n16961
|
| 51 | c2fptA_ |
|
not modelled |
77.4 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydroorotate dehydrogenase, mitochondrial;
PDBTitle: dual binding mode of a novel series of dhodh inhibitors
|
| 52 | c3ivuB_ |
|
not modelled |
76.6 |
10 |
PDB header:transferase
Chain: B: PDB Molecule:homocitrate synthase, mitochondrial;
PDBTitle: homocitrate synthase lys4 bound to 2-og
|
| 53 | c2hmcA_ |
|
not modelled |
75.8 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: the crystal structure of dihydrodipicolinate synthase dapa from2 agrobacterium tumefaciens
|
| 54 | c3ffsC_ |
|
not modelled |
75.7 |
21 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:inosine-5-monophosphate dehydrogenase;
PDBTitle: the crystal structure of cryptosporidium parvum inosine-5'-2 monophosphate dehydrogenase
|
| 55 | c3gr7A_ |
|
not modelled |
75.5 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadph dehydrogenase;
PDBTitle: structure of oye from geobacillus kaustophilus, hexagonal2 crystal form
|
| 56 | c1tv5A_ |
|
not modelled |
74.9 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydroorotate dehydrogenase homolog, mitochondrial;
PDBTitle: plasmodium falciparum dihydroorotate dehydrogenase with a bound2 inhibitor
|
| 57 | d1tv5a1 |
|
not modelled |
74.9 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 58 | c3d0cB_ |
|
not modelled |
74.2 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 oceanobacillus iheyensis at 1.9 a resolution
|
| 59 | d2a6na1 |
|
not modelled |
73.8 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 60 | d1ep3a_ |
|
not modelled |
73.7 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 61 | c3q58A_ |
|
not modelled |
73.2 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:n-acetylmannosamine-6-phosphate 2-epimerase;
PDBTitle: structure of n-acetylmannosamine-6-phosphate epimerase from salmonella2 enterica
|
| 62 | c2rfgB_ |
|
not modelled |
72.9 |
22 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from hahella2 chejuensis at 1.5a resolution
|
| 63 | c3eb2A_ |
|
not modelled |
72.8 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:putative dihydrodipicolinate synthetase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 2.0a resolution
|
| 64 | c3gnnA_ |
|
not modelled |
72.2 |
9 |
PDB header:transferase
Chain: A: PDB Molecule:nicotinate-nucleotide pyrophosphorylase;
PDBTitle: crystal structure of nicotinate-nucleotide2 pyrophosphorylase from burkholderi pseudomallei
|
| 65 | c3fkkA_ |
|
not modelled |
71.6 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:l-2-keto-3-deoxyarabonate dehydratase;
PDBTitle: structure of l-2-keto-3-deoxyarabonate dehydratase
|
| 66 | c2hl7A_ |
|
not modelled |
71.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome c-type biogenesis protein ccmh;
PDBTitle: crystal structure of the periplasmic domain of ccmh from pseudomonas2 aeruginosa
|
| 67 | c1qpoA_ |
|
not modelled |
71.0 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:quinolinate acid phosphoribosyl transferase;
PDBTitle: quinolinate phosphoribosyl transferase (qaprtase) apo-enzyme from2 mycobacterium tuberculosis
|
| 68 | c3inpA_ |
|
not modelled |
70.7 |
13 |
PDB header:isomerase
Chain: A: PDB Molecule:d-ribulose-phosphate 3-epimerase;
PDBTitle: 2.05 angstrom resolution crystal structure of d-ribulose-phosphate 3-2 epimerase from francisella tularensis.
|
| 69 | c3h5dD_ |
|
not modelled |
70.3 |
19 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: dihydrodipicolinate synthase from drug-resistant streptococcus2 pneumoniae
|
| 70 | d1xcfa_ |
|
not modelled |
70.2 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 71 | c2v9dB_ |
|
not modelled |
69.4 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:yage;
PDBTitle: crystal structure of yage, a prophage protein belonging to2 the dihydrodipicolinic acid synthase family from e. coli3 k12
|
| 72 | c3noeA_ |
|
not modelled |
68.4 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from pseudomonas2 aeruginosa
|
| 73 | d1rd5a_ |
|
not modelled |
68.2 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 74 | c3e96B_ |
|
not modelled |
68.2 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 bacillus clausii
|
| 75 | c3qfeB_ |
|
not modelled |
68.2 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:putative dihydrodipicolinate synthase family protein;
PDBTitle: crystal structures of a putative dihydrodipicolinate synthase family2 protein from coccidioides immitis
|
| 76 | c3b4uB_ |
|
not modelled |
68.1 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from agrobacterium2 tumefaciens str. c58
|
| 77 | c1zfjA_ |
|
not modelled |
67.5 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine monophosphate dehydrogenase;
PDBTitle: inosine monophosphate dehydrogenase (impdh; ec 1.1.1.205) from2 streptococcus pyogenes
|
| 78 | c2ekcA_ |
|
not modelled |
66.6 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: structural study of project id aq_1548 from aquifex aeolicus vf5
|
| 79 | c2r8wB_ |
|
not modelled |
66.3 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:agr_c_1641p;
PDBTitle: the crystal structure of dihydrodipicolinate synthase (atu0899) from2 agrobacterium tumefaciens str. c58
|
| 80 | c2yxgD_ |
|
not modelled |
65.1 |
17 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihyrodipicolinate synthase (dapa)
|
| 81 | d1vyra_ |
|
not modelled |
65.0 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 82 | c3g0sA_ |
|
not modelled |
64.9 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: dihydrodipicolinate synthase from salmonella typhimurium lt2
|
| 83 | c2jbmA_ |
|
not modelled |
64.4 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:nicotinate-nucleotide pyrophosphorylase;
PDBTitle: qprtase structure from human
|
| 84 | c2c3zA_ |
|
not modelled |
64.3 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:indole-3-glycerol phosphate synthase;
PDBTitle: crystal structure of a truncated variant of indole-3-2 glycerol phosphate synthase from sulfolobus solfataricus
|
| 85 | c3daqB_ |
|
not modelled |
63.8 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from methicillin-2 resistant staphylococcus aureus
|
| 86 | d1qopa_ |
|
not modelled |
63.4 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 87 | d1hl2a_ |
|
not modelled |
63.4 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 88 | c3s5oA_ |
|
not modelled |
62.9 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:4-hydroxy-2-oxoglutarate aldolase, mitochondrial;
PDBTitle: crystal structure of human 4-hydroxy-2-oxoglutarate aldolase bound to2 pyruvate
|
| 89 | c3bi8A_ |
|
not modelled |
62.0 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: structure of dihydrodipicolinate synthase from clostridium2 botulinum
|
| 90 | d1f76a_ |
|
not modelled |
61.4 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 91 | c1vrdA_ |
|
not modelled |
61.4 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine-5'-monophosphate dehydrogenase;
PDBTitle: crystal structure of inosine-5'-monophosphate dehydrogenase (tm1347)2 from thermotoga maritima at 2.18 a resolution
|
| 92 | d1xi3a_ |
|
not modelled |
61.3 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Thiamin phosphate synthase
Family:Thiamin phosphate synthase |
| 93 | c1o4uA_ |
|
not modelled |
61.3 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:type ii quinolic acid phosphoribosyltransferase;
PDBTitle: crystal structure of a nicotinate nucleotide pyrophosphorylase2 (tm1645) from thermotoga maritima at 2.50 a resolution
|
| 94 | c3igsB_ |
|
not modelled |
60.1 |
12 |
PDB header:isomerase
Chain: B: PDB Molecule:n-acetylmannosamine-6-phosphate 2-epimerase 2;
PDBTitle: structure of the salmonella enterica n-acetylmannosamine-6-phosphate2 2-epimerase
|
| 95 | c2kw0A_ |
|
not modelled |
59.5 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ccmh protein;
PDBTitle: solution structure of n-terminal domain of ccmh from escherichia.coli
|
| 96 | d1kbla1 |
|
not modelled |
59.1 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate phosphate dikinase, C-terminal domain |
| 97 | c3thaB_ |
|
not modelled |
59.0 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: tryptophan synthase subunit alpha from campylobacter jejuni.
|
| 98 | c1me9A_ |
|
not modelled |
58.8 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine-5'-monophosphate dehydrogenase;
PDBTitle: inosine monophosphate dehydrogenase (impdh) from2 tritrichomonas foetus with imp bound
|
| 99 | c2b7pA_ |
|
not modelled |
58.1 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:probable nicotinate-nucleotide pyrophosphorylase;
PDBTitle: crystal structure of quinolinic acid phosphoribosyltransferase from2 helicobacter pylori
|
| 100 | d1xm3a_ |
|
not modelled |
57.7 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:ThiG-like
Family:ThiG-like |
| 101 | d1ps9a1 |
|
not modelled |
55.3 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 102 | d1ujpa_ |
|
not modelled |
55.2 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 103 | c2vefB_ |
|
not modelled |
54.7 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:dihydropteroate synthase;
PDBTitle: dihydropteroate synthase from streptococcus pneumoniae
|
| 104 | c1keeH_ |
|
not modelled |
54.2 |
18 |
PDB header:ligase
Chain: H: PDB Molecule:carbamoyl-phosphate synthetase small chain;
PDBTitle: inactivation of the amidotransferase activity of carbamoyl phosphate2 synthetase by the antibiotic acivicin
|
| 105 | c2nx9B_ |
|
not modelled |
54.2 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:oxaloacetate decarboxylase 2, subunit alpha;
PDBTitle: crystal structure of the carboxyltransferase domain of the2 oxaloacetate decarboxylase na+ pump from vibrio cholerae
|
| 106 | c3n2xB_ |
|
not modelled |
54.0 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:uncharacterized protein yage;
PDBTitle: crystal structure of yage, a prophage protein belonging to the2 dihydrodipicolinic acid synthase family from e. coli k12 in complex3 with pyruvate
|
| 107 | c3lerA_ |
|
not modelled |
53.3 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 campylobacter jejuni subsp. jejuni nctc 11168
|
| 108 | c2a7nA_ |
|
not modelled |
52.9 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l(+)-mandelate dehydrogenase;
PDBTitle: crystal structure of the g81a mutant of the active chimera of (s)-2 mandelate dehydrogenase
|
| 109 | d1piia2 |
|
not modelled |
52.8 |
6 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 110 | c1jcnA_ |
|
not modelled |
51.8 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine monophosphate dehydrogenase i;
PDBTitle: binary complex of human type-i inosine monophosphate dehydrogenase2 with 6-cl-imp
|
| 111 | c3tdmD_ |
|
not modelled |
50.9 |
23 |
PDB header:de novo protein
Chain: D: PDB Molecule:computationally designed two-fold symmetric tim-barrel
PDBTitle: computationally designed tim-barrel protein, halfflr
|
| 112 | c3khjE_ |
|
not modelled |
50.0 |
20 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:inosine-5-monophosphate dehydrogenase;
PDBTitle: c. parvum inosine monophosphate dehydrogenase bound by inhibitor c64
|
| 113 | c3tr9A_ |
|
not modelled |
49.4 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: structure of a dihydropteroate synthase (folp) in complex with pteroic2 acid from coxiella burnetii
|
| 114 | d1vhna_ |
|
not modelled |
49.2 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 115 | d1xkya1 |
|
not modelled |
49.2 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 116 | d1f74a_ |
|
not modelled |
49.0 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 117 | c2nuxB_ |
|
not modelled |
48.6 |
23 |
PDB header:lyase
Chain: B: PDB Molecule:2-keto-3-deoxygluconate/2-keto-3-deoxy-6-phospho gluconate
PDBTitle: 2-keto-3-deoxygluconate aldolase from sulfolobus acidocaldarius,2 native structure in p6522 at 2.5 a resolution
|
| 118 | c2yzrB_ |
|
not modelled |
47.7 |
24 |
PDB header:lyase
Chain: B: PDB Molecule:pyridoxal biosynthesis lyase pdxs;
PDBTitle: crystal structure of pyridoxine biosynthesis protein from2 methanocaldococcus jannaschii
|
| 119 | c3dz1A_ |
|
not modelled |
47.7 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 1.87a resolution
|
| 120 | d2flia1 |
|
not modelled |
46.2 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |