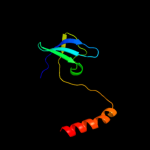
| 1 | c3d3rA_ |
|
 |
100.0 |
32 |
PDB header:chaperone
Chain: A: PDB Molecule:hydrogenase assembly chaperone hypc/hupf;
PDBTitle: crystal structure of the hydrogenase assembly chaperone hypc/hupf2 family protein from shewanella oneidensis mr-1
|
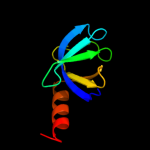
| 2 | d2ot2a1 |
|
 |
99.9 |
57 |
Fold:OB-fold
Superfamily:HupF/HypC-like
Family:HupF/HypC-like |
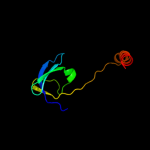
| 3 | d3d3ra1 |
|
 |
99.9 |
32 |
Fold:OB-fold
Superfamily:HupF/HypC-like
Family:HupF/HypC-like |
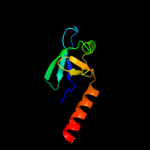
| 4 | d2z1ca1 |
|
 |
99.9 |
38 |
Fold:OB-fold
Superfamily:HupF/HypC-like
Family:HupF/HypC-like |
| 5 | d1rl2a2 |
|
 |
80.6 |
28 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 6 | c2rcnA_ |
|
 |
67.5 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:probable gtpase engc;
PDBTitle: crystal structure of the ribosomal interacting gtpase yjeq from the2 enterobacterial species salmonella typhimurium.
|
| 7 | d1vqoa2 |
|
 |
66.7 |
24 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 8 | c4a2iV_ |
|
 |
65.4 |
26 |
PDB header:ribosome/hydrolase
Chain: V: PDB Molecule:putative ribosome biogenesis gtpase rsga;
PDBTitle: cryo-electron microscopy structure of the 30s subunit in complex with2 the yjeq biogenesis factor
|
| 9 | d2zjra2 |
|
 |
56.1 |
34 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 10 | c1u0lB_ |
|
 |
55.1 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:probable gtpase engc;
PDBTitle: crystal structure of yjeq from thermotoga maritima
|
| 11 | c2dgyA_ |
|
 |
54.3 |
16 |
PDB header:translation
Chain: A: PDB Molecule:mgc11102 protein;
PDBTitle: solution structure of the eukaryotic initiation factor 1a2 in mgc11102 protein
|
| 12 | c2yv5A_ |
|
 |
54.1 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:yjeq protein;
PDBTitle: crystal structure of yjeq from aquifex aeolicus
|
| 13 | c3mxnB_ |
|
 |
53.7 |
18 |
PDB header:replication
Chain: B: PDB Molecule:recq-mediated genome instability protein 2;
PDBTitle: crystal structure of the rmi core complex
|
| 14 | c1g0dA_ |
|
 |
43.4 |
33 |
PDB header:transferase
Chain: A: PDB Molecule:protein-glutamine gamma-glutamyltransferase;
PDBTitle: crystal structure of red sea bream transglutaminase
|
| 15 | c1kv3F_ |
|
 |
42.5 |
44 |
PDB header:transferase
Chain: F: PDB Molecule:protein-glutamine gamma-glutamyltransferase;
PDBTitle: human tissue transglutaminase in gdp bound form
|
| 16 | c1l9mB_ |
|
 |
42.3 |
33 |
PDB header:transferase
Chain: B: PDB Molecule:protein-glutamine glutamyltransferase e3;
PDBTitle: three-dimensional structure of the human transglutaminase 32 enzyme: binding of calcium ions change structure for3 activation
|
| 17 | c1f13A_ |
|
 |
40.0 |
33 |
PDB header:coagulation factor
Chain: A: PDB Molecule:cellular coagulation factor xiii zymogen;
PDBTitle: recombinant human cellular coagulation factor xiii
|
| 18 | c2oqkA_ |
|
 |
39.7 |
34 |
PDB header:translation
Chain: A: PDB Molecule:putative translation initiation factor eif-1a;
PDBTitle: crystal structure of putative cryptosporidium parvum translation2 initiation factor eif-1a
|
| 19 | d2j01d2 |
|
 |
37.2 |
27 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 20 | d1ny4a_ |
|
 |
35.6 |
17 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 21 | d2qamc2 |
|
not modelled |
35.4 |
22 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 22 | d1jt8a_ |
|
not modelled |
34.9 |
16 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 23 | d1g0da4 |
|
not modelled |
33.7 |
30 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:Transglutaminase core |
| 24 | d1d7qa_ |
|
not modelled |
33.3 |
19 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 25 | d1njjb1 |
|
not modelled |
33.1 |
33 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Eukaryotic ODC-like |
| 26 | d2q3za4 |
|
not modelled |
32.8 |
44 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:Transglutaminase core |
| 27 | d1ex0a4 |
|
not modelled |
31.3 |
30 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:Transglutaminase core |
| 28 | c1t9hA_ |
|
not modelled |
31.2 |
9 |
PDB header:hydrolase
Chain: A: PDB Molecule:probable gtpase engc;
PDBTitle: the crystal structure of yloq, a circularly permuted gtpase.
|
| 29 | d1vjja4 |
|
not modelled |
29.6 |
30 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:Transglutaminase core |
| 30 | c2xzm1_ |
|
not modelled |
29.1 |
19 |
PDB header:ribosome
Chain: 1: PDB Molecule:ribosomal protein s28e containing protein;
PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
|
| 31 | d2toda1 |
|
not modelled |
28.4 |
33 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Eukaryotic ODC-like |
| 32 | c1rl2A_ |
|
not modelled |
28.3 |
17 |
PDB header:ribosomal protein
Chain: A: PDB Molecule:protein (ribosomal protein l2);
PDBTitle: ribosomal protein l2 rna-binding domain from bacillus2 stearothermophilus
|
| 33 | d1d7ka1 |
|
not modelled |
27.1 |
33 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Eukaryotic ODC-like |
| 34 | d1ne3a_ |
|
not modelled |
26.7 |
18 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 35 | c1njjC_ |
|
not modelled |
23.3 |
33 |
PDB header:lyase
Chain: C: PDB Molecule:ornithine decarboxylase;
PDBTitle: crystal structure determination of t. brucei ornithine2 decarboxylase bound to d-ornithine and to g418
|
| 36 | c3i4oA_ |
|
not modelled |
22.8 |
21 |
PDB header:translation
Chain: A: PDB Molecule:translation initiation factor if-1;
PDBTitle: crystal structure of translation initiation factor 1 from2 mycobacterium tuberculosis
|
| 37 | c3fp9E_ |
|
not modelled |
22.8 |
24 |
PDB header:hydrolase
Chain: E: PDB Molecule:proteasome-associated atpase;
PDBTitle: crystal structure of intern domain of proteasome-associated2 atpase, mycobacterium tuberculosis
|
| 38 | d1f3ta1 |
|
not modelled |
22.3 |
33 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Eukaryotic ODC-like |
| 39 | c2hc8A_ |
|
not modelled |
22.2 |
24 |
PDB header:transport protein
Chain: A: PDB Molecule:cation-transporting atpase, p-type;
PDBTitle: structure of the a. fulgidus copa a-domain
|
| 40 | c1oy5B_ |
|
not modelled |
20.2 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:trna (guanine-n(1)-)-methyltransferase;
PDBTitle: crystal structure of trna (m1g37) methyltransferase from aquifex2 aeolicus
|
| 41 | d1u0la1 |
|
not modelled |
19.7 |
18 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 42 | d1knwa1 |
|
not modelled |
19.5 |
25 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Eukaryotic ODC-like |
| 43 | c2qghA_ |
|
not modelled |
19.5 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:diaminopimelate decarboxylase;
PDBTitle: crystal structure of diaminopimelate decarboxylase from helicobacter2 pylori complexed with l-lysine
|
| 44 | d1t9ha1 |
|
not modelled |
19.1 |
14 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 45 | d7odca1 |
|
not modelled |
18.6 |
33 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Eukaryotic ODC-like |
| 46 | d1oy5a_ |
|
not modelled |
18.3 |
22 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:tRNA(m1G37)-methyltransferase TrmD |
| 47 | c1xniI_ |
|
not modelled |
17.8 |
19 |
PDB header:cell cycle
Chain: I: PDB Molecule:tumor suppressor p53-binding protein 1;
PDBTitle: tandem tudor domain of 53bp1
|
| 48 | d1twia1 |
|
not modelled |
17.8 |
8 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Eukaryotic ODC-like |
| 49 | c2p3eA_ |
|
not modelled |
16.9 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:diaminopimelate decarboxylase;
PDBTitle: crystal structure of aq1208 from aquifex aeolicus
|
| 50 | c1kqsA_ |
|
not modelled |
16.5 |
24 |
PDB header:ribosome
Chain: A: PDB Molecule:ribosomal protein l2;
PDBTitle: the haloarcula marismortui 50s complexed with a2 pretranslocational intermediate in protein synthesis
|
| 51 | c2nvaH_ |
|
not modelled |
15.5 |
33 |
PDB header:lyase
Chain: H: PDB Molecule:arginine decarboxylase, a207r protein;
PDBTitle: the x-ray crystal structure of the paramecium bursaria2 chlorella virus arginine decarboxylase bound to agmatine
|
| 52 | c1knwA_ |
|
not modelled |
14.9 |
25 |
PDB header:lyase
Chain: A: PDB Molecule:diaminopimelate decarboxylase;
PDBTitle: crystal structure of diaminopimelate decarboxylase
|
| 53 | d3pnpa_ |
|
not modelled |
14.4 |
22 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 54 | c2yxxA_ |
|
not modelled |
14.3 |
25 |
PDB header:lyase
Chain: A: PDB Molecule:diaminopimelate decarboxylase;
PDBTitle: crystal structure analysis of diaminopimelate decarboxylate (lysa)
|
| 55 | c1yr3A_ |
|
not modelled |
13.7 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:xanthosine phosphorylase;
PDBTitle: escherichia coli purine nucleoside phosphorylase ii, the2 product of the xapa gene
|
| 56 | d1p3ha_ |
|
not modelled |
13.6 |
28 |
Fold:GroES-like
Superfamily:GroES-like
Family:GroES |
| 57 | d1p3qq_ |
|
not modelled |
13.5 |
33 |
Fold:RuvA C-terminal domain-like
Superfamily:UBA-like
Family:CUE domain |
| 58 | c1w4sA_ |
|
not modelled |
13.4 |
38 |
PDB header:nuclear protein
Chain: A: PDB Molecule:polybromo 1 protein;
PDBTitle: crystal structure of the proximal bah domain of polybromo
|
| 59 | d1aono_ |
|
not modelled |
13.3 |
24 |
Fold:GroES-like
Superfamily:GroES-like
Family:GroES |
| 60 | d1cb0a_ |
|
not modelled |
13.2 |
11 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 61 | d1fr3a_ |
|
not modelled |
12.9 |
11 |
Fold:OB-fold
Superfamily:MOP-like
Family:Molybdate/tungstate binding protein MOP |
| 62 | d2id1a1 |
|
not modelled |
12.8 |
27 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:Iojap/YbeB-like |
| 63 | d1uala_ |
|
not modelled |
12.7 |
38 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:tRNA(m1G37)-methyltransferase TrmD |
| 64 | d2a7ya1 |
|
not modelled |
12.6 |
22 |
Fold:SH3-like barrel
Superfamily:Cell growth inhibitor/plasmid maintenance toxic component
Family:Rv2302-like |
| 65 | c2a7yA_ |
|
not modelled |
12.6 |
22 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein rv2302/mt2359;
PDBTitle: solution structure of the conserved hypothetical protein2 rv2302 from the bacterium mycobacterium tuberculosis
|
| 66 | d1vmka_ |
|
not modelled |
12.5 |
30 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 67 | c2akwB_ |
|
not modelled |
12.0 |
15 |
PDB header:ligase
Chain: B: PDB Molecule:phenylalanyl-trna synthetase beta chain;
PDBTitle: crystal structure of t.thermophilus phenylalanyl-trna synthetase2 complexed with p-cl-phenylalanine
|
| 68 | c2pljA_ |
|
not modelled |
11.7 |
25 |
PDB header:lyase
Chain: A: PDB Molecule:lysine/ornithine decarboxylase;
PDBTitle: crystal structure of lysine/ornithine decarboxylase complexed with2 putrescine from vibrio vulnificus
|
| 69 | c2k29A_ |
|
not modelled |
11.5 |
25 |
PDB header:transcription
Chain: A: PDB Molecule:antitoxin relb;
PDBTitle: structure of the dbd domain of e. coli antitoxin relb
|
| 70 | d1ntga_ |
|
not modelled |
11.4 |
17 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Myf domain |
| 71 | d1pqwa_ |
|
not modelled |
11.3 |
31 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Alcohol dehydrogenase-like, C-terminal domain |
| 72 | c3n29A_ |
|
not modelled |
11.2 |
8 |
PDB header:lyase
Chain: A: PDB Molecule:carboxynorspermidine decarboxylase;
PDBTitle: crystal structure of carboxynorspermidine decarboxylase complexed with2 norspermidine from campylobacter jejuni
|
| 73 | c3knuD_ |
|
not modelled |
11.2 |
33 |
PDB header:transferase
Chain: D: PDB Molecule:trna (guanine-n(1)-)-methyltransferase;
PDBTitle: crystal structure of trna (guanine-n1)-methyltransferase from2 anaplasma phagocytophilum
|
| 74 | d1g2oa_ |
|
not modelled |
11.1 |
22 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 75 | c3upsA_ |
|
not modelled |
11.0 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:iojap-like protein;
PDBTitle: crystal structure of iojap-like protein from zymomonas mobilis
|
| 76 | c3quvB_ |
|
not modelled |
10.9 |
33 |
PDB header:transferase
Chain: B: PDB Molecule:trna (guanine-n(1)-)-methyltransferase;
PDBTitle: crystal structure of a trna-guanine-n1-methyltransferase from2 mycobacterium abscessus
|
| 77 | c3khsB_ |
|
not modelled |
10.8 |
26 |
PDB header:hydrolase
Chain: B: PDB Molecule:purine nucleoside phosphorylase;
PDBTitle: crystal structure of grouper iridovirus purine nucleoside2 phosphorylase
|
| 78 | d1v3va2 |
|
not modelled |
10.8 |
31 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Alcohol dehydrogenase-like, C-terminal domain |
| 79 | c2j66A_ |
|
not modelled |
10.7 |
45 |
PDB header:lyase
Chain: A: PDB Molecule:btrk;
PDBTitle: structural characterisation of btrk decarboxylase from2 butirosin biosynthesis
|
| 80 | c3ky7A_ |
|
not modelled |
10.7 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:trna (guanine-n(1)-)-methyltransferase;
PDBTitle: 2.35 angstrom resolution crystal structure of a putative trna2 (guanine-7-)-methyltransferase (trmd) from staphylococcus aureus3 subsp. aureus mrsa252
|
| 81 | c3e6eC_ |
|
not modelled |
10.6 |
15 |
PDB header:isomerase
Chain: C: PDB Molecule:alanine racemase;
PDBTitle: crystal structure of alanine racemase from e.faecalis2 complex with cycloserine
|
| 82 | d2fzwa2 |
|
not modelled |
10.6 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Alcohol dehydrogenase-like, C-terminal domain |
| 83 | d1p9pa_ |
|
not modelled |
10.5 |
33 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:tRNA(m1G37)-methyltransferase TrmD |
| 84 | c1ltlE_ |
|
not modelled |
10.4 |
22 |
PDB header:replication
Chain: E: PDB Molecule:dna replication initiator (cdc21/cdc54);
PDBTitle: the dodecamer structure of mcm from archaeal m.2 thermoautotrophicum
|
| 85 | c2equA_ |
|
not modelled |
10.4 |
17 |
PDB header:protein binding
Chain: A: PDB Molecule:phd finger protein 20-like 1;
PDBTitle: solution structure of the tudor domain of phd finger2 protein 20-like 1
|
| 86 | c1tufA_ |
|
not modelled |
10.4 |
8 |
PDB header:lyase
Chain: A: PDB Molecule:diaminopimelate decarboxylase;
PDBTitle: crystal structure of diaminopimelate decarboxylase from m.2 jannaschi
|
| 87 | c3iefA_ |
|
not modelled |
10.3 |
22 |
PDB header:transferase, rna binding protein
Chain: A: PDB Molecule:trna (guanine-n(1)-)-methyltransferase;
PDBTitle: crystal structure of trna guanine-n1-methyltransferase from2 bartonella henselae using mpcs.
|
| 88 | c3ggsA_ |
|
not modelled |
10.2 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:purine nucleoside phosphorylase;
PDBTitle: human purine nucleoside phosphorylase double mutant e201q,n243d2 complexed with 2-fluoro-2'-deoxyadenosine
|
| 89 | c3mt1B_ |
|
not modelled |
10.2 |
8 |
PDB header:lyase
Chain: B: PDB Molecule:putative carboxynorspermidine decarboxylase protein;
PDBTitle: crystal structure of putative carboxynorspermidine decarboxylase2 protein from sinorhizobium meliloti
|
| 90 | c2p4sA_ |
|
not modelled |
10.1 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:purine nucleoside phosphorylase;
PDBTitle: structure of purine nucleoside phosphorylase from anopheles gambiae in2 complex with dadme-immh
|
| 91 | d2fsqa1 |
|
not modelled |
10.0 |
6 |
Fold:LigT-like
Superfamily:LigT-like
Family:Atu0111-like |
| 92 | d2hd3a1 |
|
not modelled |
10.0 |
33 |
Fold:OB-fold
Superfamily:EutN/CcmL-like
Family:EutN/CcmL-like |
| 93 | d2qw7a1 |
|
not modelled |
9.9 |
24 |
Fold:OB-fold
Superfamily:EutN/CcmL-like
Family:EutN/CcmL-like |
| 94 | c3oo2A_ |
|
not modelled |
9.8 |
16 |
PDB header:isomerase
Chain: A: PDB Molecule:alanine racemase 1;
PDBTitle: 2.37 angstrom resolution crystal structure of an alanine racemase2 (alr) from staphylococcus aureus subsp. aureus col
|
| 95 | d2soba_ |
|
not modelled |
9.7 |
9 |
Fold:OB-fold
Superfamily:Staphylococcal nuclease
Family:Staphylococcal nuclease |
| 96 | d1ltla_ |
|
not modelled |
9.6 |
21 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:DNA replication initiator (cdc21/cdc54) N-terminal domain |
| 97 | d2nn6h1 |
|
not modelled |
9.5 |
16 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 98 | c2opkC_ |
|
not modelled |
9.4 |
27 |
PDB header:isomerase
Chain: C: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of a putative mannose-6-phosphate isomerase2 (reut_a1446) from ralstonia eutropha jmp134 at 2.10 a resolution
|
| 99 | c2zkra_ |
|
not modelled |
9.4 |
25 |
PDB header:ribosomal protein/rna
Chain: A: PDB Molecule:rna expansion segment es3;
PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
|

