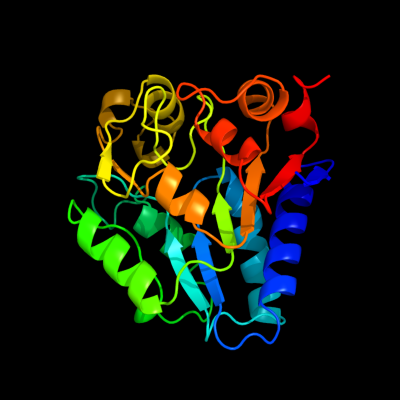
| 1 | d1ne7a_
|
|
 |
100.0 |
29 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:NagB-like |
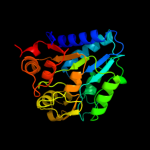
| 2 | c2bkxB_
|
|
 |
100.0 |
27 |
PDB header:hydrolase
Chain: B: PDB Molecule:glucosamine-6-phosphate deaminase;
PDBTitle: structure and kinetics of a monomeric glucosamine-6-2 phosphate deaminase: missing link of the nagb superfamily
|
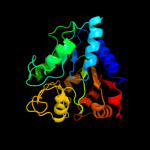
| 3 | c3hn6D_
|
|
 |
100.0 |
32 |
PDB header:isomerase
Chain: D: PDB Molecule:glucosamine-6-phosphate deaminase;
PDBTitle: crystal structure of glucosamine-6-phosphate deaminase from borrelia2 burgdorferi
|
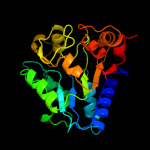
| 4 | d1fsfa_
|
|
 |
100.0 |
28 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:NagB-like |
| 5 | c2ri0B_
|
|
 |
100.0 |
27 |
PDB header:hydrolase
Chain: B: PDB Molecule:glucosamine-6-phosphate deaminase;
PDBTitle: crystal structure of glucosamine 6-phosphate deaminase (nagb) from s.2 mutans
|
| 6 | c3icoA_
|
|
 |
100.0 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:6-phosphogluconolactonase;
PDBTitle: crystal structure of 6-phosphogluconolactonase from2 mycobacterium tuberculosis
|
| 7 | c2j0eA_
|
|
 |
100.0 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:6-phosphogluconolactonase;
PDBTitle: three dimensional structure and catalytic mechanism of 6-2 phosphogluconolactonase from trypanosoma brucei
|
| 8 | c3oc6A_
|
|
 |
100.0 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:6-phosphogluconolactonase;
PDBTitle: crystal structure of 6-phosphogluconolactonase from mycobacterium2 smegmatis, apo form
|
| 9 | c1y89B_
|
|
 |
100.0 |
23 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:devb protein;
PDBTitle: crystal structure of devb protein
|
| 10 | c3e15D_
|
|
 |
100.0 |
17 |
PDB header:hydrolase
Chain: D: PDB Molecule:glucose-6-phosphate 1-dehydrogenase;
PDBTitle: 6-phosphogluconolactonase from plasmodium vivax
|
| 11 | c3cssA_
|
|
 |
100.0 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:6-phosphogluconolactonase;
PDBTitle: crystal structure of 6-phosphogluconolactonase from leishmania2 guyanensis
|
| 12 | d1vl1a_
|
|
 |
100.0 |
19 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:NagB-like |
| 13 | c1pbtA_
|
|
 |
100.0 |
19 |
PDB header:hydrolase, oxidoreductase
Chain: A: PDB Molecule:6-phosphogluconolactonase;
PDBTitle: the crystal structure of tm1154, oxidoreductase, sol/devb2 family from thermotoga maritima
|
| 14 | c3lhiA_
|
|
 |
100.0 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative 6-phosphogluconolactonase;
PDBTitle: crystal structure of putative 6-2 phosphogluconolactonase(yp_207848.1) from neisseria3 gonorrhoeae fa 1090 at 1.33 a resolution
|
| 15 | c3lwdA_
|
|
 |
100.0 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:6-phosphogluconolactonase;
PDBTitle: crystal structure of putative 6-phosphogluconolactonase (yp_574786.1)2 from chromohalobacter salexigens dsm 3043 at 1.88 a resolution
|
| 16 | c3nwpA_
|
|
 |
100.0 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:6-phosphogluconolactonase;
PDBTitle: crystal structure of a 6-phosphogluconolactonase (sbal_2240) from2 shewanella baltica os155 at 1.40 a resolution
|
| 17 | d2gnpa1
|
|
 |
100.0 |
12 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:SorC sugar-binding domain-like |
| 18 | c2w48D_
|
|
 |
100.0 |
11 |
PDB header:transcription
Chain: D: PDB Molecule:sorbitol operon regulator;
PDBTitle: crystal structure of the full-length sorbitol operon2 regulator sorc from klebsiella pneumoniae
|
| 19 | c3nzeB_
|
|
 |
100.0 |
15 |
PDB header:transcription regulator
Chain: B: PDB Molecule:putative transcriptional regulator, sugar-binding family;
PDBTitle: the crystal structure of a domain of a possible sugar-binding2 transcriptional regulator from arthrobacter aurescens tc1.
|
| 20 | c3kv1A_
|
|
 |
100.0 |
11 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional repressor;
PDBTitle: crystal structure of putative sugar-binding domain of transcriptional2 repressor from vibrio fischeri
|
| 21 | c2o0mA_ |
|
not modelled |
100.0 |
15 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator, sorc family;
PDBTitle: the crystal structure of the putative sorc family transcriptional2 regulator from enterococcus faecalis
|
| 22 | d2o0ma1 |
|
not modelled |
100.0 |
15 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:SorC sugar-binding domain-like |
| 23 | d2okga1 |
|
not modelled |
100.0 |
17 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:SorC sugar-binding domain-like |
| 24 | d3efba1 |
|
not modelled |
100.0 |
12 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:SorC sugar-binding domain-like |
| 25 | d2r5fa1 |
|
not modelled |
100.0 |
22 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:SorC sugar-binding domain-like |
| 26 | c3u7jA_ |
|
not modelled |
95.5 |
13 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: crystal structure of ribose-5-phosphate isomerase a from burkholderia2 thailandensis
|
| 27 | c3hheA_ |
|
not modelled |
93.3 |
21 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: crystal structure of ribose-5-phosphate isomerase a from bartonella2 henselae
|
| 28 | c1uj6A_ |
|
not modelled |
91.0 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose 5-phosphate isomerase;
PDBTitle: crystal structure of thermus thermophilus ribose-5-phosphate isomerase2 complexed with arabinose-5-phosphate
|
| 29 | c1lk5C_ |
|
not modelled |
90.8 |
18 |
PDB header:isomerase
Chain: C: PDB Molecule:d-ribose-5-phosphate isomerase;
PDBTitle: structure of the d-ribose-5-phosphate isomerase from2 pyrococcus horikoshii
|
| 30 | d1uj4a1 |
|
not modelled |
89.4 |
15 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:D-ribose-5-phosphate isomerase (RpiA), catalytic domain |
| 31 | c2f8mB_ |
|
not modelled |
89.0 |
15 |
PDB header:isomerase
Chain: B: PDB Molecule:ribose 5-phosphate isomerase;
PDBTitle: ribose 5-phosphate isomerase from plasmodium falciparum
|
| 32 | c1xtzA_ |
|
not modelled |
89.0 |
19 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose-5-phosphate isomerase;
PDBTitle: crystal structure of the s. cerevisiae d-ribose-5-phosphate isomerase:2 comparison with the archeal and bacterial enzymes
|
| 33 | c1m0sA_ |
|
not modelled |
87.8 |
30 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: northeast structural genomics consortium (nesg id ir21)
|
| 34 | c2pjmA_ |
|
not modelled |
87.6 |
10 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: structure of ribose 5-phosphate isomerase a from2 methanocaldococcus jannaschii
|
| 35 | c3kwmC_ |
|
not modelled |
86.3 |
19 |
PDB header:isomerase
Chain: C: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: crystal structure of ribose-5-isomerase a
|
| 36 | c3l7oB_ |
|
not modelled |
85.9 |
14 |
PDB header:isomerase
Chain: B: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: crystal structure of ribose-5-phosphate isomerase a from streptococcus2 mutans ua159
|
| 37 | c1lkzB_ |
|
not modelled |
85.7 |
17 |
PDB header:isomerase
Chain: B: PDB Molecule:ribose 5-phosphate isomerase a;
PDBTitle: crystal structure of d-ribose-5-phosphate isomerase (rpia)2 from escherichia coli.
|
| 38 | d1lk5a1 |
|
not modelled |
83.5 |
20 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:D-ribose-5-phosphate isomerase (RpiA), catalytic domain |
| 39 | d1m0sa1 |
|
not modelled |
80.7 |
30 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:D-ribose-5-phosphate isomerase (RpiA), catalytic domain |
| 40 | d1o8bb1 |
|
not modelled |
78.1 |
19 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:D-ribose-5-phosphate isomerase (RpiA), catalytic domain |
| 41 | c2i2aA_ |
|
not modelled |
59.7 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:probable inorganic polyphosphate/atp-nad kinase 1;
PDBTitle: crystal structure of lmnadk1 from listeria monocytogenes
|
| 42 | d1ni5a1 |
|
not modelled |
59.2 |
8 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:PP-loop ATPase |
| 43 | d1wxia1 |
|
not modelled |
54.7 |
19 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:N-type ATP pyrophosphatases |
| 44 | c1ni5A_ |
|
not modelled |
37.8 |
8 |
PDB header:cell cycle
Chain: A: PDB Molecule:putative cell cycle protein mesj;
PDBTitle: structure of the mesj pp-atpase from escherichia coli
|
| 45 | d1xnga1 |
|
not modelled |
36.8 |
14 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:N-type ATP pyrophosphatases |
| 46 | c3p52B_ |
|
not modelled |
35.8 |
12 |
PDB header:ligase
Chain: B: PDB Molecule:nh(3)-dependent nad(+) synthetase;
PDBTitle: nh3-dependent nad synthetase from campylobacter jejuni subsp. jejuni2 nctc 11168 in complex with the nitrate ion
|
| 47 | c3tqcB_ |
|
not modelled |
32.2 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:pantothenate kinase;
PDBTitle: structure of the pantothenate kinase (coaa) from coxiella burnetii
|
| 48 | d1zuna1 |
|
not modelled |
31.2 |
7 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:PAPS reductase-like |
| 49 | c1zunA_ |
|
not modelled |
31.1 |
7 |
PDB header:transferase
Chain: A: PDB Molecule:sulfate adenylyltransferase subunit 2;
PDBTitle: crystal structure of a gtp-regulated atp sulfurylase2 heterodimer from pseudomonas syringae
|
| 50 | c2hfqA_ |
|
not modelled |
28.0 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein;
PDBTitle: nmr structure of protein ne1680 from nitrosomonas europaea:2 northeast structural genomics consortium target net5
|
| 51 | d2hfqa1 |
|
not modelled |
28.0 |
14 |
Fold:NE1680-like
Superfamily:NE1680-like
Family:NE1680-like |
| 52 | d1kqpa_ |
|
not modelled |
25.0 |
23 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:N-type ATP pyrophosphatases |
| 53 | c3dpiA_ |
|
not modelled |
24.6 |
20 |
PDB header:ligase
Chain: A: PDB Molecule:nad+ synthetase;
PDBTitle: crystal structure of nad+ synthetase from burkholderia pseudomallei
|
| 54 | c2gaaA_ |
|
not modelled |
24.1 |
23 |
PDB header:unknown function
Chain: A: PDB Molecule:hypothetical 39.9 kda protein;
PDBTitle: crystal structure of yfh7 from saccharomyces cerevisiae: a2 putative p-loop containing kinase with a circular3 permutation.
|
| 55 | c2c40B_ |
|
not modelled |
20.2 |
5 |
PDB header:hydrolase
Chain: B: PDB Molecule:inosine-uridine preferring nucleoside hydrolase family
PDBTitle: crystal structure of inosine-uridine preferring nucleoside2 hydrolase from bacillus anthracis at 2.2a resolution
|
| 56 | c3clhA_ |
|
not modelled |
19.7 |
9 |
PDB header:lyase
Chain: A: PDB Molecule:3-dehydroquinate synthase;
PDBTitle: crystal structure of 3-dehydroquinate synthase (dhqs)from2 helicobacter pylori
|
| 57 | c1xahA_ |
|
not modelled |
19.2 |
6 |
PDB header:lyase
Chain: A: PDB Molecule:3-dehydroquinate synthase;
PDBTitle: crystal structure of staphlyococcus aureus 3-dehydroquinate2 synthase (dhqs) in complex with zn2+ and nad+
|
| 58 | c1yqtA_ |
|
not modelled |
18.5 |
17 |
PDB header:hydrolyase/translation
Chain: A: PDB Molecule:rnase l inhibitor;
PDBTitle: rnase-l inhibitor
|
| 59 | d1wy5a1 |
|
not modelled |
17.6 |
13 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:PP-loop ATPase |
| 60 | c3fiuD_ |
|
not modelled |
17.6 |
10 |
PDB header:ligase
Chain: D: PDB Molecule:nh(3)-dependent nad(+) synthetase;
PDBTitle: structure of nmn synthetase from francisella tularensis
|
| 61 | c3c7tB_ |
|
not modelled |
17.4 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:ecdysteroid-phosphate phosphatase;
PDBTitle: crystal structure of the ecdysone phosphate phosphatase, eppase, from2 bombix mori in complex with tungstate
|
| 62 | d1xhoa_ |
|
not modelled |
16.6 |
9 |
Fold:Bacillus chorismate mutase-like
Superfamily:YjgF-like
Family:Chorismate mutase |
| 63 | d3bzka5 |
|
not modelled |
15.7 |
17 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Tex RuvX-like domain-like |
| 64 | c3ozxA_ |
|
not modelled |
15.6 |
17 |
PDB header:hydrolase, translation
Chain: A: PDB Molecule:rnase l inhibitor;
PDBTitle: crystal structure of abce1 of sulfolubus solfataricus (-fes domain)
|
| 65 | c3hjgB_ |
|
not modelled |
14.8 |
12 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative alpha-ribazole-5'-phosphate phosphatase
PDBTitle: crystal structure of putative alpha-ribazole-5'-phosphate2 phosphatase cobc from vibrio parahaemolyticus
|
| 66 | d1v43a3 |
|
not modelled |
14.8 |
14 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:ABC transporter ATPase domain-like |
| 67 | c3q4gA_ |
|
not modelled |
14.2 |
20 |
PDB header:ligase
Chain: A: PDB Molecule:nh(3)-dependent nad(+) synthetase;
PDBTitle: structure of nad synthetase from vibrio cholerae
|
| 68 | d1pjqa3 |
|
not modelled |
13.9 |
19 |
Fold:Siroheme synthase middle domains-like
Superfamily:Siroheme synthase middle domains-like
Family:Siroheme synthase middle domains-like |
| 69 | c2p4sA_ |
|
not modelled |
13.8 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:purine nucleoside phosphorylase;
PDBTitle: structure of purine nucleoside phosphorylase from anopheles gambiae in2 complex with dadme-immh
|
| 70 | c3ju3A_ |
|
not modelled |
13.2 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable 2-oxoacid ferredoxin oxidoreductase, alpha chain;
PDBTitle: crystal structure of alpha chain of probable 2-oxoacid ferredoxin2 oxidoreductase from thermoplasma acidophilum
|
| 71 | d1ujna_ |
|
not modelled |
13.0 |
9 |
Fold:Dehydroquinate synthase-like
Superfamily:Dehydroquinate synthase-like
Family:Dehydroquinate synthase, DHQS |
| 72 | d1k6da_ |
|
not modelled |
12.3 |
15 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:CoA transferase alpha subunit-like |
| 73 | c2gruB_ |
|
not modelled |
12.0 |
6 |
PDB header:lyase
Chain: B: PDB Molecule:2-deoxy-scyllo-inosose synthase;
PDBTitle: crystal structure of 2-deoxy-scyllo-inosose synthase2 complexed with carbaglucose-6-phosphate, nad+ and co2+
|
| 74 | d2hhja1 |
|
not modelled |
11.5 |
10 |
Fold:Phosphoglycerate mutase-like
Superfamily:Phosphoglycerate mutase-like
Family:Cofactor-dependent phosphoglycerate mutase |
| 75 | c2ghiD_ |
|
not modelled |
11.5 |
22 |
PDB header:transport protein
Chain: D: PDB Molecule:transport protein;
PDBTitle: crystal structure of plasmodium yoelii multidrug resistance2 protein 2
|
| 76 | d3dhwc1 |
|
not modelled |
11.4 |
17 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:ABC transporter ATPase domain-like |
| 77 | c3dqqB_ |
|
not modelled |
11.3 |
11 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative trna synthase;
PDBTitle: the crystal structure of the putative trna synthase from salmonella2 typhimurium lt2
|
| 78 | c3dl2A_ |
|
not modelled |
11.1 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:ubiquitin-conjugating enzyme e2 variant 3;
PDBTitle: hexagonal structure of the ldh domain of human ubiquitin-2 conjugating enzyme e2-like isoform a
|
| 79 | c3d4iD_ |
|
not modelled |
11.0 |
15 |
PDB header:hydrolase
Chain: D: PDB Molecule:sts-2 protein;
PDBTitle: crystal structure of the 2h-phosphatase domain of sts-2
|
| 80 | d1k6ma2 |
|
not modelled |
10.7 |
13 |
Fold:Phosphoglycerate mutase-like
Superfamily:Phosphoglycerate mutase-like
Family:6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase, phosphatase domain |
| 81 | c3okfA_ |
|
not modelled |
10.6 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:3-dehydroquinate synthase;
PDBTitle: 2.5 angstrom resolution crystal structure of 3-dehydroquinate synthase2 (arob) from vibrio cholerae
|
| 82 | c3bk7A_ |
|
not modelled |
10.6 |
21 |
PDB header:hydrolyase/translation
Chain: A: PDB Molecule:abc transporter atp-binding protein;
PDBTitle: structure of the complete abce1/rnaase-l inhibitor protein2 from pyrococcus abysii
|
| 83 | d1bifa2 |
|
not modelled |
10.4 |
13 |
Fold:Phosphoglycerate mutase-like
Superfamily:Phosphoglycerate mutase-like
Family:6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase, phosphatase domain |
| 84 | d3d31a2 |
|
not modelled |
10.3 |
17 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:ABC transporter ATPase domain-like |
| 85 | d1dbfa_ |
|
not modelled |
10.2 |
12 |
Fold:Bacillus chorismate mutase-like
Superfamily:YjgF-like
Family:Chorismate mutase |
| 86 | c2pcjB_ |
|
not modelled |
10.2 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:lipoprotein-releasing system atp-binding protein lold;
PDBTitle: crystal structure of abc transporter (aq_297) from aquifex aeolicus2 vf5
|
| 87 | d1g2912 |
|
not modelled |
9.9 |
17 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:ABC transporter ATPase domain-like |
| 88 | c3fvqB_ |
|
not modelled |
9.9 |
23 |
PDB header:hydrolase
Chain: B: PDB Molecule:fe(3+) ions import atp-binding protein fbpc;
PDBTitle: crystal structure of the nucleotide binding domain fbpc2 complexed with atp
|
| 89 | c2i55C_ |
|
not modelled |
9.7 |
18 |
PDB header:isomerase
Chain: C: PDB Molecule:phosphomannomutase;
PDBTitle: complex of glucose-1,6-bisphosphate with phosphomannomutase from2 leishmania mexicana
|
| 90 | d1oxxk2 |
|
not modelled |
9.6 |
21 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:ABC transporter ATPase domain-like |
| 91 | d2gnoa2 |
|
not modelled |
9.2 |
18 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Extended AAA-ATPase domain |
| 92 | d1gpma1 |
|
not modelled |
9.2 |
15 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:N-type ATP pyrophosphatases |
| 93 | c2e21A_ |
|
not modelled |
8.8 |
14 |
PDB header:ligase
Chain: A: PDB Molecule:trna(ile)-lysidine synthase;
PDBTitle: crystal structure of tils in a complex with amppnp from aquifex2 aeolicus.
|
| 94 | c3fzqA_ |
|
not modelled |
8.7 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative hydrolase;
PDBTitle: crystal structure of putative haloacid dehalogenase-like hydrolase2 (yp_001086940.1) from clostridium difficile 630 at 2.10 a resolution
|
| 95 | c1tcvB_ |
|
not modelled |
8.6 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:purine-nucleoside phosphorylase;
PDBTitle: crystal structure of the purine nucleoside phosphorylase2 from schistosoma mansoni in complex with non-detergent3 sulfobetaine 195 and acetate
|
| 96 | c3c6kC_ |
|
not modelled |
8.5 |
16 |
PDB header:transferase
Chain: C: PDB Molecule:spermine synthase;
PDBTitle: crystal structure of human spermine synthase in complex2 with spermidine and 5-methylthioadenosine
|
| 97 | d1sgwa_ |
|
not modelled |
8.5 |
17 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:ABC transporter ATPase domain-like |
| 98 | c2cbzA_ |
|
not modelled |
8.2 |
14 |
PDB header:transport
Chain: A: PDB Molecule:multidrug resistance-associated protein 1;
PDBTitle: structure of the human multidrug resistance protein 12 nucleotide binding domain 1
|
| 99 | d2g39a2 |
|
not modelled |
8.1 |
11 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:CoA transferase alpha subunit-like |